I have two sigmoid functions, sig1 and sig2, made with this function sigmoid = function(x, A =1, mu=0, ss = 1) A*1 / (1 + exp(-(x-mu) * ss)). Since they have a true offset of 10 (mu1=50 and mu2=60) I expected their cross-correlation function to peak at a lag of 10. My non-mathematical intuition is that cross-correlation "slides" one of the curves over by a given lag, correlates, and repeats for multiple lags. When I do this by myself in code (my.ccf in the code below) I do directly recover the lags I designed into the curves. However, ccf, the real R cross-correlation function, returns max lag of 4 (see below). What's going on?
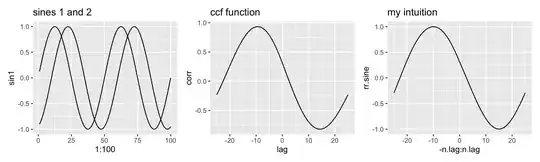
To replicate, I did the same thing for two sines. They have a designed "lag" of 10 (see below). Here, the max lag returned by ccf (9) is closer to the designed-in value, but isn't exactly equal. My intuitive function returns 10, the "correct" answer.
Why doesn't the max of the two cross-correlation functions (4 and 9) exactly equal the lag I coded into the curves (10 and 10, respectively)? What's wrong with my intuition? Edit: As pointed out by Whuber, why isn't the max of ccf 1, since these are identical and perfectly aligned vectors.
(code)
# sigmoid function
sigmoid = function(x, A =1, mu=0, ss = 1) A*1 / (1 + exp(-(x-mu) * ss))
# my intuition
my.ccf = function(x,y, lag=20) {
lags = -lag : lag
# add padding to y
y.padded = c(rep(NA,lag), y, rep(NA,lag))
# correlate
rr = numeric(length(lags))
for (ii in 1:length(lags)) {
# apply lag to y.padded
I = (1:length(x)) + (ii-1)
y.lagged = y.padded[I]
rr[ii] = cor.test(x, y.lagged)$estimate
}
return(rr)
}
# make sigmoids and cross-correlate
sig1 = sigmoid(1:100, mu=50, ss=1/3)
sig2 = sigmoid(1:100, mu=60, ss=1/3)
ccf.sig=ccf(sig1, sig2, plot=F)
rr.sig = my.ccf(sig2,sig1,lag=16)
# do the same with sines
sin1 = sin((1:100) * 4*pi/100)
sin2 = sin(((1:100) - 10) * 4*pi/100)
ccf.sine=ccf(sin1, sin2,plot=F)
rr.sine = my.ccf(sin2,sin1,lag=16)
# plot sigmoids + ccf.sig
p1 = ggplot() + geom_line( aes(x=1:100,y=sig1)) +
geom_line(aes(x=1:100,y=sig2)) + ggtitle("sigmoids 1 and 2")
p2 = ggplot(data.frame(lag=ccf.sig$lag, corr=ccf.sig$acf), aes(x=lag, y=corr)) +
geom_line() + ggtitle("ccf function")
p3 = ggplot() + geom_line(aes(x=-16:16, y=rr.sine)) + ggtitle("my intuition")
p1 + p2 + p3
# plot sines + ccf.sig
p1 = ggplot() + geom_line( aes(x=1:100,y=sin1)) +
geom_line(aes(x=1:100,y=sin2)) + ggtitle("sines 1 and 2")
p2 = ggplot(data.frame(lag=ccf.sine$lag, corr=ccf.sine$acf), aes(x=lag, y=corr)) +
geom_line() + ggtitle("ccf function")
p3 = ggplot() + geom_line(aes(x=-16:16, y=rr.sig)) + ggtitle("my intuition")
p1 + p2 + p3