I'm trying to estimate using MLE the parameters of a (Gamma) probability distribution given some data, but the data contain a good number of samples below the minimum detection limit (MDL) of the measurement method. These show up as zeroes in the dataset. I came up with a piecewise likelihood function where all 0s are assigned the mean probability density over [0,MDL], i.e. CDF(MDL)/MDL. And then all values above the MDL just have their likelihood calculated as normal.
Questions:
- Is there anything WRONG with this approach? Are there better approaches?
- Are there any non-obvious situations where it will fail to reliably estimate the parameters?
- Has this been done in the literature before? If so, does somebody have a citation I can use?
Reprex below...
x_actual <- rgamma(100, 1,1)
MDL <- .25
x_with_0s <- x_actual
x_with_0s[x_actual<MDL] <- 0
likfunc <- function(k, theta){
mean_lik_below_MDL <- pgamma(MDL, k, theta)/MDL
liks <- log(dgamma(x, k, theta))
liks[x==0] <- log(mean_lik_below_MDL)
loglik <- -sum(liks)
return(loglik)
}
This seemingly returns pretty good parameter estimates:
> x <- x_actual
> mle(likfunc, start=list(k=.5, theta=.5), method = "L-BFGS-B", lower = c(0,0))
Call:
mle(minuslogl = likfunc, start = list(k = 0.5, theta = 0.5),
method = "L-BFGS-B", lower = c(0, 0))
Coefficients:
k theta
1.082315 1.077583
> x <- x_with_0s
> mle(likfunc, start=list(k=.5, theta=.5), method = "L-BFGS-B", lower = c(0,0))
Call:
mle(minuslogl = likfunc, start = list(k = 0.5, theta = 0.5),
method = "L-BFGS-B", lower = c(0, 0))
Coefficients:
k theta
1.031814 1.029893
>
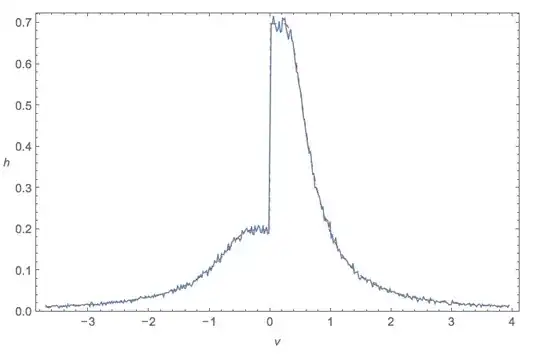
One thing I don't like is that there's a discontinuity in the PDF for the likelihood function, but I don't know if this is really an issue.
x <- seq(0,5, .01)
pdf <- function(k, theta){
mean_lik_below_MDL <- pgamma(MDL, k, theta)/MDL
liks <- dgamma(x, k, theta)
liks[x<MDL] <- mean_lik_below_MDL
return(liks)
}
plot(pdf(1,1)~x)