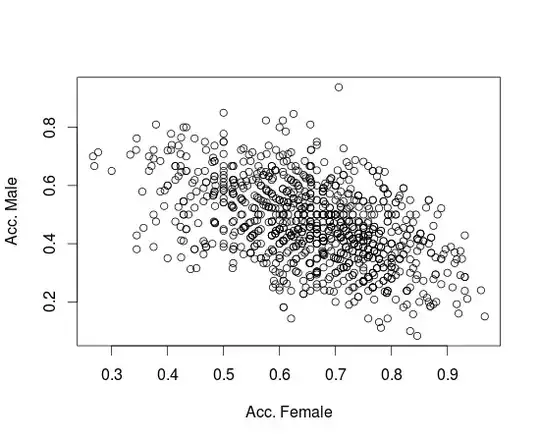
I have a typical binary classification problem with a sample of ~700 instances where I fitted multiple classification models including logistic regression, SVM and Random Forest.
The instances are represented by legacy features, and the classification results have been unsatisfying. The information retreival measures obtained on the test dataset vary significantly depending on the used model. Precision is in the range of 0.7-0.9 and recall is in the range of 0.5-0.7.
I was trying to informally argue that there is a drift in the population of positive instances. Both my sample and previous samples are biased so I cannot compare features computed on both.
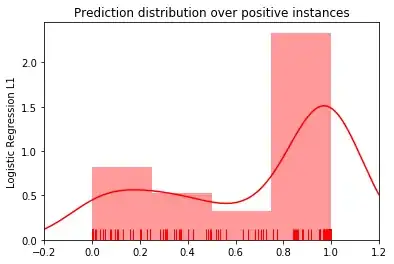
Instead, I am arguing that the legacy features are not effective for the classification task, which is concurred by the classification results. I was exploring the possibility of illustrating this point visually and plotted the distribution of predictions probabilities for positive and negative instances (prediction probability for each instance computed using the scikit-learn predict_proba).
Here is the distribution for the SVM model. It seems that there is a bimodal distribution of the prediction probabilities.
Same for Logistic Regression.
Random Forest on the other hand gives me a unimodal distribution:
I have repeated the above for different sets of legacy features and obtained similar results. The mode positions change but I always have a bi- or tri-modal distributions for all models except Random Forest.
I am not sure how to robustly interpret these plots. It seems to me that there are two (or more) distinct populations of positive instances and that the legacy features are effective for one of these and not useful with the other(s).
I have no explanations for the Random Forest distribution, however. And I do not even know where to look for explanations. Does it have anything to do with Random Forest being an ensemble classifier? And if so what exactly?
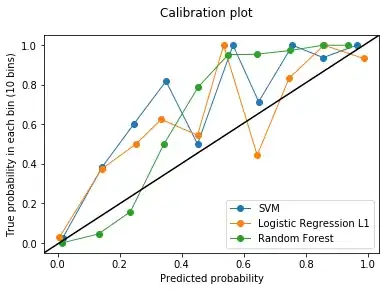
Edit (07/02/2020): Adding the calibration curves in response to the comment by usεr11852 below.