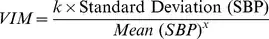Although I find this reading regarding the interpretation of the Coefficent of Variation (CoV) very interesting, I did not found hints regarding the Variability Independent of the Mean (VIM) which is beeing largely employed in cardiovascular research. In particular, VIM is calculated by: $$VIM=(SD/mean^x)$$
fitting a curve through a plot of SD systolic BP (SBP; y axis) against mean SBP (x axis) with the parameter x estimated from the curve
Moreover, I found other different definitions/approaches:
Computation 2:
VIM is calculated as the SD divided by the mean to the power
xand multiplied by the population mean to the powerx. The powerxis obtained by fitting a curve through a plot of SD against mean using the modelSD= a × mean x, wherexwas derived by nonlinear regression analysis as implemented in thePROC NLINprocedure of the SAS package.
Computation 3:
VIM is calculated as
100 * SD/mean^β, whereβis the regression coefficient based on natural logarithm of SD on natural logarithm of mean
Computation 4:
VIM is a transformation of the standard deviation that is uncorrelated with mean BP and is calculated as follows:
Where x is calculated from fitting a power model:
(i.e. the mean of all SBP readings across all patients to the power of x).
Herein I propose a reproducible R example of computing SD and CoV. I would like to add the VIM too:
set.seed(101)
df <- data.frame( "ID" = paste("ID", seq(1,100,1), sep = ""),
"x1" = sample(90:220, size = 100, replace = T),
"x2" = sample(90:220, size = 100, replace = T),
"x3" = sample(90:220, size = 100, replace = T),
"x4" = sample(90:220, size = 100, replace = T),
"x5" = sample(90:220, size = 100, replace = T))
# Compute row average
df$avg <- round(rowMeans(df[2:6]), 1)
# Compute row standard deviation
df$sntd <- round(matrixStats::rowSds(as.matrix(df[2:6])), 1)
# Compute coefficient of variation (CoV) as SD/mean * 100
df$CoV <- round(df$sntd/df$avg*100, 1)
To the best of my understanding, probably nsl R function can be used for non-linear regression. How?
Can anyone help with VIM computing using R and suggest/explain the above mentioned VIM definitions?