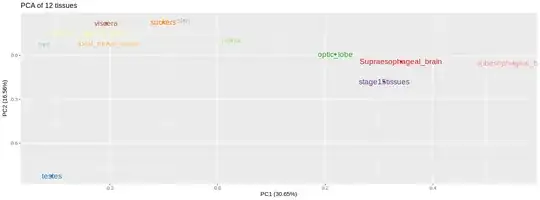
I'm analyzing the expression of a dataset of genes in 12 tissues(the rownames). I constructed a matrix with genes as rows and tissues as columns. I transposed the matrix to observe the PCA of tissues. I would to know the similarity among tissues. can anyone interpret these pca scores?
pca$x
PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8 PC9 PC10
ova -94.33510 16.182389 24.4399504 -58.339345 4.875150 -28.908618 25.6659749 -14.2054007 -56.436362 -16.793651
testes -90.18614 -177.378225 3.9908015 40.064548 2.181639 7.954307 -4.0899375 -3.0380629 3.031315 1.169147
retina 7.39437 22.177408 15.4855311 -20.098193 29.324046 113.730363 14.3130541 10.6977148 -5.380255 -2.930546
optic_lobe 64.02055 1.369854 50.5034514 -32.590520 26.378548 -9.078160 -52.2296341 -19.6505681 6.171714 47.686948
subesophageal_brain 160.96257 -11.183784 55.3184079 52.167708 -32.687533 -13.319774 54.1822785 -0.9204015 -14.343083 14.356696
Supraesophageal_brain 99.74678 -9.140050 40.5914715 -5.559214 15.613576 -16.787563 -37.4422182 9.1914754 10.078844 -64.486786
axial_nerve_corde -59.22247 17.585141 44.8949023 -35.908376 12.447542 -23.555745 0.9355398 2.3417200 4.288873 15.527077
suckers -29.57641 48.697031 -50.2427241 72.453724 39.101050 -20.835103 -16.0832215 57.8530926 -22.278607 11.660217
skin -18.35160 51.419553 -63.6591345 58.871896 32.202296 -2.237463 6.5965222 -69.1374409 12.025851 -8.929713
stage15tissues 90.55248 -38.593376 -137.6867820 -72.184864 -19.372106 -7.428407 5.8128683 8.0449506 1.052072 5.693779
posterior_salivary_gland -70.47519 31.835133 17.3374690 -20.468310 -3.191976 -19.863079 38.2413639 21.3112057 64.542026 -1.769445
viscera -60.52984 47.028927 -0.9733445 21.590946 -106.872232 20.329241 -35.9025905 -2.4882850 -2.752390 -1.183724
PC11 PC12
ova 19.8191933 2.670267e-13
testes 1.4740740 -6.435220e-13
retina -1.1163530 1.386079e-13
optic_lobe 21.0661258 -3.045990e-14
subesophageal_brain -1.9034947 2.533877e-13
Supraesophageal_brain -2.5045387 1.709133e-13
axial_nerve_corde -52.9084858 6.080764e-14
suckers 3.4646382 -3.608751e-14
skin -3.6858974 2.900367e-13
stage15tissues -4.5598460 -5.765999e-13
posterior_salivary_gland 20.6050331 -1.400667e-14
viscera 0.2495509 1.529362e-13
I plotted the pca using :
ggplot2::autoplot(pca, label= TRUE, colour= col.pal.paired)