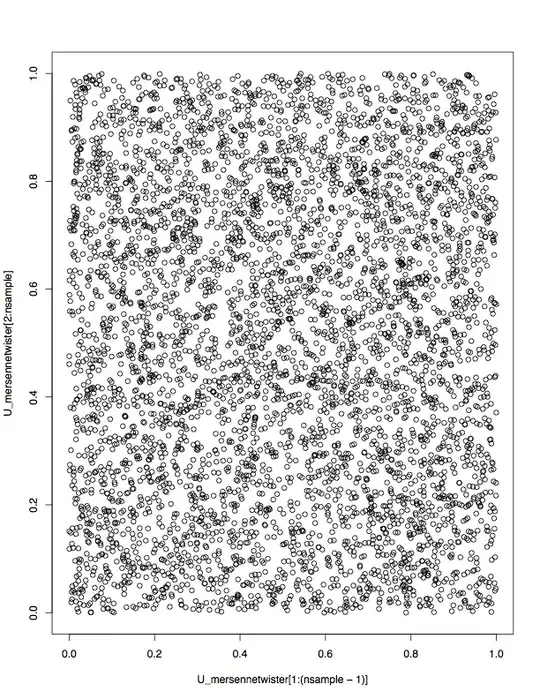
pseudo random number generators should give as output random sequences u1, u2, ... that are independent and identically distribuited (iid). Since testing for independence is not easy, the first check is testing for noncorrelation. As a first visual test you could check the scatterplot of (Ui, Ui-1).
Let's do that for the R built in function runif (Mersenne Twister generator):
nsample <- 5000 #number of random uniform numbers that I'll generate
set.seed(111) #set the seed
U_mersennetwister <- runif(nsample) #generate nsample pseudo RN using the built-in R function (ie Mersenne Twister algorithm)
plot(U_mersennetwister[1:(nsample-1)],U_mersennetwister[2:nsample]) #scatterplot (Ui vs Ui-1)
The output, as expected span the unit square almost evenly, so we can conclude that Ui is incorrelated to Ui-1:
However, if I repeat the same procedure for the built-in function rnorm, I get that the following output, which not span evenly the two axis, so for example the probability that Zi-1 < 0 given that Zi = 3 is much smaller than the probability that Zi-1 < 0 given that Zi = 0:
N <- rnorm(100000)
plot(N[1:99999],N[2:100000])
This is the output:
Can someone explain me why (Zi, Zi-1) does not span evenly the two axis?