When you fit a generalized linear model, you must specify two things. First you specify a distribution family* that describes how the variance of outcome values changes with their mean. Second, you specify a link function that describes how the linear-predictor values from the model correspond to mean outcome values.
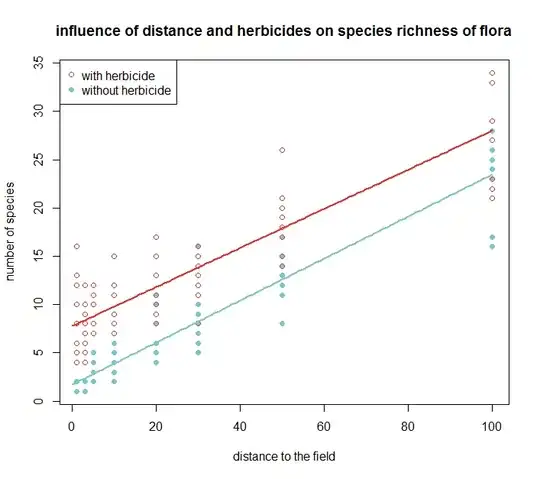
For count values, the Poisson family is a standard initial choice; the binomial family is generally reserved for yes/no or success/failure types of outcomes. For the Poisson family the variance equals the mean. That said, i'm a little concerned about whether your data will be well represented by the Poisson, as you have no 0 values even at low mean response counts. For example, the two "without herbicide" sets of cases with smallest "distance to the field" values have mean counts of about 1.5. A Poisson distribution with a mean value of 1.5 should have 0 counts in about 22% of cases, so you might need to consider an alternative.
The fitted regression model provides a combined linear predictor value for any specified values of your predictor variables. For example, say that your model has an intercept $\beta_0$ for the case without herbicide and 0 "distance to the field," a slope $\beta_1$ with respect to "distance to the field," and a fixed effect $\beta_2$ of "herbicide" independent of "distance to the field" (no interaction). Call "distance to the field" $d$ and the "herbicide" predictor $h$ with a value of 0/1 without/with herbicide. The the linear predictor for any specified $d$ and $h$ values would be $\beta_0 + \beta_1 d + \beta_2 h$. The link function is chosen to map the linear-predictor values to predicted mean values of responses. According to your plot, it looks like the mean observed counts change directly with respect to such a linear predictor, so a choice of an identity link rather than the R default log link could be reasonable.
As with any regression, it is important to check how well the model fits the data and to amend the model accordingly. That might end up requiring different choices of distribution family and link function.
I think that both @kjetil b halvorsen and I have some additional concerns about your specific experimental design, depending on which there might be some additional terms required in the formulation of your model, perhaps including a random effect, to account for potential correlations among observations. The above, however, should help clarify your choices of distribution family and link function in any event.
*You call this a "correction" in your question. Again, it helps to use the agreed-on terminology. Visiting this site can help you learn the terminology if your courses are somewhat deficient in that respect.