I have a response variable that is bimodal (basically, 2 normal distributions that are sticked together) and want to model it using a linear mixed effect model.
Here is a quick example (in R):
library(mixtools)
n1 =500
n2 =500
x = rnorm(n1,mean = 10)
y = rnorm(n2,mean = 15)
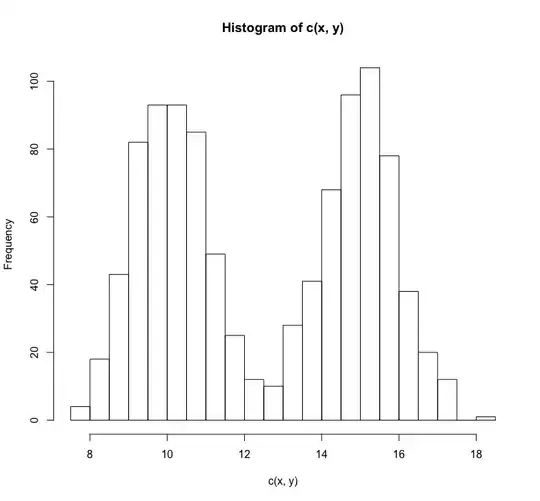
hist(c(x,y),breaks =25)
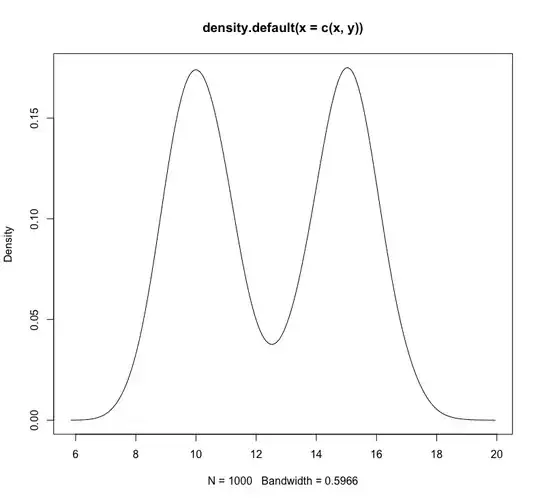
plot(density(c(x,y)))
I can run an Expectation-Maximization algorithm for gaussian mixture to get the two distributions (this is a very simple example so the 2 distributions cluster very well)
ores = mixtools::normalmixEM(c(x,y),
sigma = NULL,
mean.constr = NULL,
sd.constr = NULL,
epsilon = 1e-15,
maxit = 1000,
maxrestarts=50,
# verb = TRUE,
fast=FALSE,
ECM = FALSE,
arbmean = TRUE,
arbvar = TRUE)
ores
plot(ores,whichplots = 2)
My question is:
- Is it possible to model this bimodal variable as a response variable in a linear mixed effect model (or a GLMM if there exists a link function for that)?
- Should I need to separate the bimodal distribution in 2 distinct unimodal Gaussian distributions and construct 2 identical models but using each distribution in the separate models?
- What would be the effect of modelling a bimodal distribution with a linear mixed effect model (with a unimodal residual error)?
Finally, I heard that quantile normalization would be a way to compare the 2 distributions. How can quantile normalization be used to compare the 2 distributions in a linear mixed effect model?