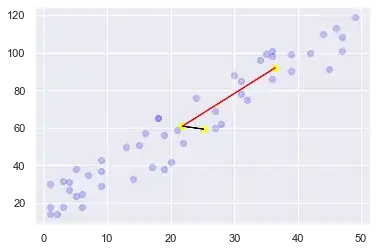
I was curious about the relationship between eigenvalues/vectors and least squares regression as well. I made widget in Sage (free math software based on python) to explore the question. The code is at the bottom of this post.
Below is a screenshot of the widget. The purple line is the regression line, the blue lines are the eigenvectors multiplied by their eigenvalues. The data points are normalized (subtracted mean, divided by standard deviation).
From the screenshot or from playing around with the values, it's clear that the eigenvectors in most cases don't point in the same direction as the regression line.
However, there is a connection between the eigenvalues and the regression line (when the data is standardized): the largest eigenvalue is equal to 1 + the regression coefficient, but this relationship goes away when the data is not normalized.
Perhaps someone else can explain exactly why that is :-)
You should be able to copy-paste this into the Sage notebook to make it work. Or run it online on cocalc.com.
import numpy as np
from numpy import linalg as LA
@interact
def f(gauss_sigma=slider(0.01,50,step_size=0.5, default=0.2),
slope=slider(-5,5,step_size=0.1, default=0.2),
points=slider(10,200,step_size=10, default=20),
seed=slider(10,20,step_size=1, default=10)):
l = points
sigma = gauss_sigma
T = RealDistribution('gaussian', sigma, seed = seed)
ranvals = [T.get_random_element() for i in range(l)]
xpts = list(range(l))
ypts = [slope*i+ranvals[i] for i in range(l)]
#Means
y_mean = mean(ypts)
x_mean = mean(xpts)
#Subtract means
xpts = [i-x_mean for i in xpts]
ypts = [i-y_mean for i in ypts]
#New means(=(0,0))
y_mean = mean(ypts)
x_mean = mean(xpts)
ympt = point((0, y_mean), color="red")
xmpt = point((x_mean, 0), color="red")
#Divide by standard deviation
xstd = std(xpts).n()
ystd = std(ypts).n()
xpts = [i/xstd for i in xpts]
ypts = [i/ystd for i in ypts]
pts = [[xpts[i], ypts[i]] for i in range(l)]
s = scatter_plot(pts)
#Covariance matrix
covma = np.cov(xpts,ypts)
corma = np.corrcoef(xpts,ypts)
#Eigenvalues, eigenvectors
w, v = LA.eig(covma)
eig1 = w[0]*vector(v[:,0])
eig2 = w[1]*vector(v[:,1])
eig1line = line([(x_mean,y_mean),(eig1[0]+x_mean, eig1[1]+y_mean)], zorder=10)
eig2line = line([(x_mean,y_mean),(eig2[0]+x_mean, eig2[1]+y_mean)], zorder=11)
#Regression line, OLS
var('x,b,d')
model(x) = b*x + d
regr = find_fit(pts, model)
f(x) = regr[0]*x + regr[1]
regr_line = line([(-35, f(-35).rhs()) ,(35,f(35).rhs() )], color="purple")
#Ellipses
from math import acos
ang = acos(vector(v[:,0]).dot_product(vector([1,0])))
elip = ellipse((x_mean,y_mean), w[0]*vector(v[:,0]).norm(), w[1]*vector(v[:,1]).norm(), ang, color="grey")
show(s + ympt + xmpt + eig1line + eig2line + regr_line, #+ elip,
xmin=-5, xmax=5, ymin=-5, ymax=5, aspect_ratio=1, figsize=10)
print("Covariance matrix:", covma, "\n",
"Correlation matrix.", corma, "\n",
"Eigenvalues:",w,
"Eigenvectors:", v, "\n",
"Regr coeffs:", regr, "\n",
sep="\n")