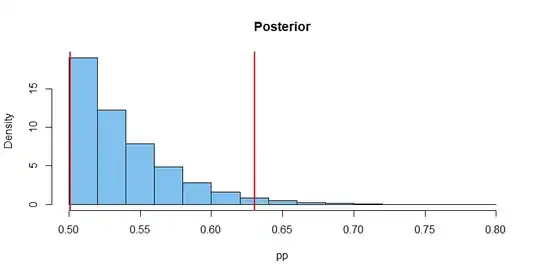
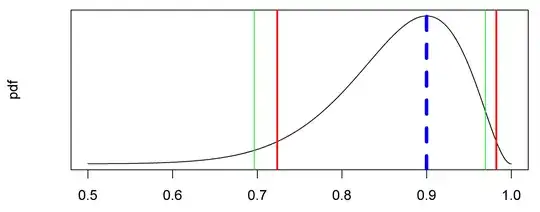
A standard problem is estimation of confidence intervals for a population proportion given that one has observed f successes out of n independent trials. There is a reasonable discussion of this problem at https://en.wikipedia.org/wiki/Binomial_proportion_confidence_interval and several R packages perform the calculation using a variety of methods (e.g. binom.confint in the package binom or bintol.int in the package interval). My problem is similar except that I know for certain that the population proportion lies in the range 0.5 to 1; within that range a uniform prior would be appropriate, although I am not wedded to a Bayesian approach. Of course particular small samples may nevertheless yield fewer successes than failures. Sample sizes are typically 10-30 trials, occasionally lower.
Ideally I would like an R function or some lines of code that would generate appropriate confidence limits. Otherwise some advice about other approaches (e.g. online calculators, programming in C) would be appreciated. My probability theory is rusty!
The motivation for this is analysing some genetic crosses following the rules of Mendelian genetics. I am crossing a heterozygote female with a homozygote recessive male and then with a homozygote dominant male. If the first male fathers all offspring, one expects a roughly 50:50 ratio of phenotypes (equal numbers of double recessives and hetoryzgotes); if the second male fathers all offspring, they will all exhibit the dominant phenotype. But sperm from both matings may be used, so any intermediate proportion between 0.5 and 1 is possible. This seems like a standard problem in genetics, which is what leads me to expect that there is a ready-made solution out there.
Many thanks!