I try to fit sin function with brms using next code:
library(tidyverse)
library(brms)
N <- 100
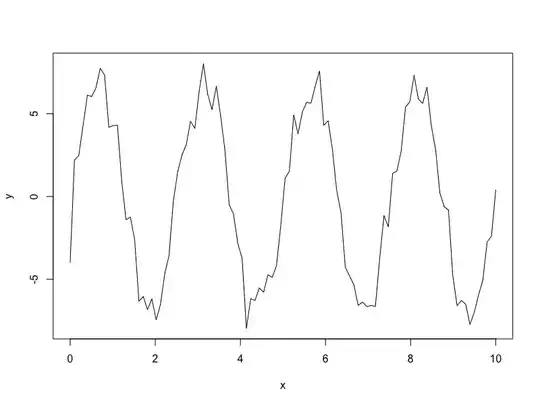
x <- seq(0, 10, length.out = N)
e <- rnorm(N)
y <- 7 * sin(2.5 * x) + e
inp_data <- tibble(x, y)
plot(inp_data, type = 'l')
priors <- prior(normal(7, 100), nlpar = "a") +
prior(normal(2.5, 100), nlpar = "b")
sin_fit <- brm(bf(y ~ a * sin(b * x), a + b ~ 1, nl = TRUE),
data = inp_data, prior = priors, refresh = 0)
The result is:
summary(sin_fit)
The model has not converged (some Rhats are > 1.1). Do not analyse the results!
We recommend running more iterations and/or setting stronger priors.
Family: gaussian
Links: mu = identity; sigma = identity
Formula: y ~ a * sin(b * x)
a ~ 1
b ~ 1
Data: inp_data (Number of observations: 100)
Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup samples = 4000
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
a_Intercept -0.00 6.83 -7.06 7.07 2 51.50
b_Intercept -0.00 2.51 -2.51 2.51 2 762.60
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
sigma 1.00 0.07 0.87 1.16 2902 1.00
Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample is
a crude measure of effective sample size, and Rhat is the potential scale
reduction factor on split chains (at convergence, Rhat = 1).
plot(sin_fit)
How to get the right values (a = 7 and b = 2.5)? What kind of prior should I use?