I have a dataset with repeated (but not always) measures of animals. I need to control for observer bias in measuring the morphology of my animal. I'm wondering what is the way to first estimate repeatability, and then correct the measurements of the animals based on the most repeatable observer.
Below, I tried to write a script that would do that, but estimating repeatability is not giving me what I expect.
In the script below, I simulated 2 people (John and Emily) measuring 20 times the same organism (10 each). I created the data so that they should be very repeatable.
#Load the package
library(rptR)
# Make fake data
animal.id = rep(LETTERS,each=10)
obs = c(rep(c("John","Emily"),130),
rep(c("John","Emily"),130))
meas = abs(rnorm(length(obs), mean = 14, sd = 1))
# put data in data.frame
data = data.frame(animal.id,obs, meas)
# Order the data to see the differences in measurement of the same animal
data = data[order(data[,c("animal.id")],data[,c("obs")]),]
# Add different values to each observer (test for differences in measuring the same animal)
data[data$obs=="John","meas"] <- data[data$obs=="John","meas"] + rnorm(26, mean = 1,sd = 0)
data[data$obs=="Emily","meas"] <-data[data$obs=="Emily","meas"] + rnorm(26, mean = 1,sd = 0)
# Define parameters
nboot = 100
npermut = 100
# Find repeatability
x1=rpt(meas ~ 1 + (1|animal.id),
grname = c("animal.id"),
data = data,
datatype ="Gaussian",
nboot = nboot,
npermut=npermut)
plot(x1)
x1
This is the result from the script above:
Repeatability estimation using the lmm method
Repeatability for animal.id
R = 0
SE = 0
CI = [0, 0]
P = 0.5 [LRT]
0.38 [Permutation]
This means that they are not repeatable. So, is there a way to find the repeatability per observer and then correct the actual measurements based on the most repeatable observer?
I guess that if someone adds always 1mm to the measured trait this could be corrected in the raw data.
Maybe I'm interpreting this wrong, and when people say that they "correct" the measurement based on repeatability, they just put a random effect of observer and it accounts for the variability that observer put in the response.
EDIT
I made a new script and found that the function ranef might be useful
# Make fake data
animal.id = c(rep(LETTERS,each=10),"C")
obs = c(rep(c("John","doh","Emily"),43),
rep(c("John","doh","Emily"),44))
# put data in data.frame
data = data.frame(animal.id,obs#, meas
)
data$meas = NA
ani.id = unique(animal.id)
for (i in 1:length(ani.id)) {
# Extract a phenotype from the population phenotype distribution
pop.dist = rnorm(n = 1,
mean = 14,
sd = 1)
rowss = which(data$animal.id == ani.id[i])
# Make a certain individual a certain value
data[rowss,"meas"] <- rnorm(n = length(rowss),
mean = pop.dist,
sd = 0)
}
# Order the data to see the differences in measurement of the same animal
data = data[order(data[,c("animal.id")], data[,c("obs")]),]
# Add different values to each observer (test for differences in measuring the same animal)
data[data$obs=="John","meas"] <- data[data$obs=="John","meas"] + rnorm(length(data[data$obs=="John","meas"]),
mean = 2,sd = 0.5)
data[data$obs=="doh","meas"] <- data[data$obs=="doh","meas"] + rnorm(length(data[data$obs=="doh","meas"]),
mean = 5,sd = 0.8)
data[data$obs=="Emily","meas"] <-data[data$obs=="Emily","meas"] + rnorm(length(data[data$obs=="Emily","meas"]),
mean = 0,sd = 0.7)
library(lme4)
x2 = lmer(meas ~ (1|obs)+ (1|animal.id), data = data )
x3 = lmer(meas ~ -1 + obs+ (1|animal.id), data = data )
x4 = lmer(meas ~ obs + (1|animal.id), data = data )
summary(x2)
x2
icc(x2)
round(fixef(x2))
round(fixef(x3))
round(fixef(x4))
fixef(x4)
ranef(x2)$obs
rr1=ranef(x2, condVar = TRUE)
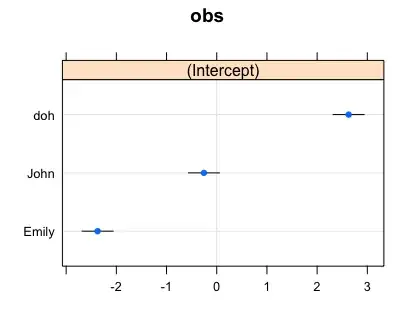
dotplot(rr1)
I think this is pretty convincing now. From model 4 lmer(meas ~ obs + (1|animal.id), data = data), the fixed effect looks like this:
round(fixef(x4))
(Intercept) obsEmily obsJohn
19 -5 -3
Here is the simulation:
John (mean = 2, sd = 0.5)
doh (mean = 5, sd = 0.8)
Emily (mean = 0, sd = 0.7)
This means that Intercept is the population mean (pop.dist = rnorm(n = 1, mean = 14, sd = 1)) plus the increase in measure that "doh" is adding (5). John compared to doh adds -3 (2 from the population mean) and Emily adds nothing (-5) compared to doh (5).
The script below calculates the repeatability of each observer.
# Define parameters
nboot = 100
npermut = 100
my.list = NULL
# Find repeatability
measurer = levels(data$obs)
for(i in 1:length(measurer)){
# How many measures were done by this guy
tmp = data[data$obs==measurer[i],]
x1=rpt(meas ~ 1 + (1|animal.id),
grname = c("animal.id"),
data = tmp,
datatype ="Gaussian",
nboot = nboot,
npermut=npermut)
plot(x1)
my.list = c(my.list,list(x1))
print(x1)
}
my.list
Here is the answer to the repeatability!
[[1]]
Repeatability estimation using the lmm method
Repeatability for animal.id
R = 0.588
SE = 0.109
CI = [0.349, 0.743]
P = 1 [LRT]
0.01 [Permutation]
[[2]]
Repeatability estimation using the lmm method
Repeatability for animal.id
R = 0.631
SE = 0.082
CI = [0.449, 0.767]
P = 1 [LRT]
0.01 [Permutation]
[[3]]
Repeatability estimation using the lmm method
Repeatability for animal.id
R = 0.819
SE = 0.051
CI = [0.668, 0.883]
P = 1 [LRT]
0.01 [Permutation]
EDIT2
I noticed that based on the information that we have on an animal, it might be better to use the most relevant biological value for the "correction" of the phenotypic measurements.
mean.not.corrected=mean(data$meas)
data$obs = factor(data$obs,levels(data$obs)[c(2,1,3)])
meas.eff = fixef(x4)
data[data$obs=="doh","meas"] = data[data$obs=="doh","meas"]-meas.eff[2]
data[data$obs=="John","meas"] = data[data$obs=="John","meas"]-meas.eff[3]
mean(data$meas)
x5 = lmer(meas ~ (1|obs)+ (1|animal.id), data = data )
x6 = lmer(meas ~ -1 + obs+ (1|animal.id), data = data )
x7 = lmer(meas ~ obs + (1|animal.id), data = data )
round(fixef(x5))
round(fixef(x6))
mean.corrected = mean(data$meas)
As you can see the corrected mean (13.7) is closer to the population mean (14) compared to the uncorrected mean is 16.1.
mean.corrected
[1] 13.709
mean.not.corrected
[1] 16.11122
Here you can see that there is no difference between the observers:
round(fixef(x7))
(Intercept) obsdoh obsJohn
14 0 0
You can now see that there is no random effect.
ranef(x5)
$obs
(Intercept)
Emily 0
doh 0
John 0
EDIT 3
# In the parametric world
hist(data$meas)
av.out1 = aov(meas ~ obs + animal.id, data=data)
av.out2 = aov(meas ~ obs, data=data)
summary(av.out1)
TukeyHSD(av.out1,"obs")
TukeyHSD(av.out2,"obs")
# In the non parametric framework, there is no difference (Mann-Whitney-Wilcoxon Test)
nonpar.data1 = data[data$obs %in% c("John","Emily") ,]
nonpar.data2 = data[data$obs %in% c("John","doh") ,]
nonpar.data3 = data[data$obs %in% c("doh","Emily") ,]
wilcox.test(meas ~ obs, data=nonpar.data1)
wilcox.test(meas ~ obs, data=nonpar.data2)
wilcox.test(meas ~ obs, data=nonpar.data3)
# There is no difference in the non-parametric Kruskal-Wallis Test
kruskal.test(meas ~ obs, data = data)
Does that make sense?