I help a colleague to bootstrap a meta-analysis mixed-effects model using the metafor R package framework authored by @Wolfgang.
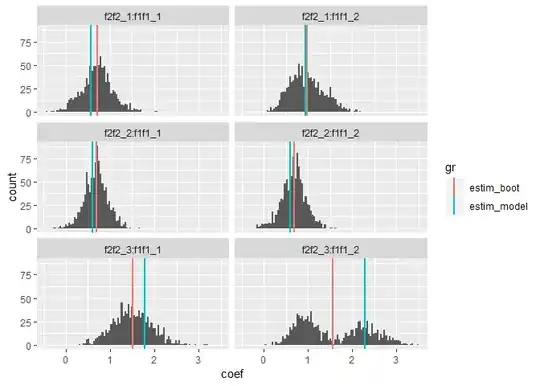
Interestingly and worryingly, for one of the model's coefficients I get a bimodal distribution when bootstrapping (see the bottom-right panel of the figure below).
I guess one of the main causes could be the fact that when bootstrapping, say half of the models converge in a local solution and the other half in another one. I tried to tune the convergence algorithm as suggested in this metafor documentation - Convergence Problems with the rma() function. Also, I tried other convergence algorithms like bobyqa and newuoa as suggested in the help documentation of rma.mv function, but got the same bimodal response.
I also tried to eliminate some of the potential outliers from the problematic group as suggested in How to interpret multimodal distribution of bootstrapped correlation, but to no avail.
I couldn't find a way to reproduce this so I uploaded data on a GitHub repository (also the links in the code section below should load in your environment all that is needed to test the case). I run the bootstrapping on a Linux cluster as an array job (just in case, the shell script is job.sh, which executes on each CPU the R script bootstrap.r that runs the model described below). A single run takes 2-3 minutes. Note that bootstrapping 100 times is also enough to detect the bimodal response. Below is an example for 1000 iterations. I am familiar with R and other methods but not that much with meta-analysis.
I would appreciate help with understanding if the bimodal distribution is ok (though might be due to convergence issues) and if not, then what can one do about it? (besides what I tried already)
Below - comparing coefficients from bootstrapping (red lines) and from a single full model run (blue lines). The histograms depict the bootstrapped distributions for each coefficient. Sampling the data for bootstrapping was done as selecting with replacement from each group/combination formed by the two fixed effects. Their raw sample sizes are:
table(dt$f1, dt$f2)
#>
#> f2_1 f2_2 f2_3
#> f1_1 177 174 41
#> f1_2 359 363 107
library(data.table)
library(ggplot2)
library(metafor)
#> Loading required package: Matrix
#> Loading 'metafor' package (version 2.0-0). For an overview
#> and introduction to the package please type: help(metafor).
load(url("https://github.com/valentinitnelav/test/raw/master/bimodal_distrib_boot/coef_boot_dt_1010.rda"))
load(url("https://github.com/valentinitnelav/test/raw/master/bimodal_distrib_boot/rmamv_model.rda"))
load(url("https://github.com/valentinitnelav/test/raw/master/bimodal_distrib_boot/data.rda"))
coef_dt <- data.frame(estim = rmamv_model[["beta"]])
coef_dt$coef_name <- rownames(coef_dt)
coef_dt <- rbind(coef_dt,
coef_boot_dt[, .(estim = mean(coef)), by = coef_name])
coef_dt[, gr := rep(c("estim_model", "estim_boot"), each = 6)]
ggplot(data = coef_boot_dt,
aes(x = coef,
group = coef_name)) +
geom_histogram(bins = 100) +
geom_vline(aes(xintercept = estim,
group = gr,
color = gr),
lwd = 1,
data = coef_dt) +
facet_wrap(vars(coef_name), ncol = 2)
Created on 2019-05-02 by the reprex package (v0.2.1)
The model goes like this:
rmamv_model <- rma.mv(y ~ f2:f1 - 1,
V = var_y,
random = list(~ 1|r1,
~ 1|r2),
R = list(r2 = cor_mat),
data = dt,
method = "REML",
# Tune the convergence algorithm / optimizer
control = list(optimizer = "nlminb",
iter.max = 1000,
step.min = 0.4,
step.max = 0.5))
R session info:
devtools::session_info()
#> - Session info ----------------------------------------------------------
#> setting value
#> version R version 3.5.2 (2018-12-20)
#> os Windows 7 x64 SP 1
#> system x86_64, mingw32
#> ui RTerm
#> language (EN)
#> collate English_United States.1252
#> ctype English_United States.1252
#> date 2019-05-02
#>
#> - Packages --------------------------------------------------------------
#> package * version date lib source
#> assertthat 0.2.1 2019-03-21 [1] CRAN (R 3.5.2)
#> backports 1.1.3 2018-12-14 [1] CRAN (R 3.5.2)
#> callr 3.2.0 2019-03-15 [1] CRAN (R 3.5.3)
#> cli 1.1.0 2019-03-19 [1] CRAN (R 3.5.3)
#> colorspace 1.4-1 2019-03-18 [1] CRAN (R 3.5.3)
#> crayon 1.3.4 2017-09-16 [1] CRAN (R 3.5.1)
#> curl 3.3 2019-01-10 [1] CRAN (R 3.5.2)
#> data.table * 1.12.0 2019-01-13 [1] CRAN (R 3.5.2)
#> desc 1.2.0 2018-05-01 [1] CRAN (R 3.5.1)
#> devtools 2.0.1 2018-10-26 [1] CRAN (R 3.5.1)
#> digest 0.6.18 2018-10-10 [1] CRAN (R 3.5.1)
#> dplyr 0.8.0.1 2019-02-15 [1] CRAN (R 3.5.2)
#> evaluate 0.13 2019-02-12 [1] CRAN (R 3.5.2)
#> fs 1.2.7 2019-03-19 [1] CRAN (R 3.5.3)
#> ggplot2 * 3.1.0 2018-10-25 [1] CRAN (R 3.5.1)
#> glue 1.3.1 2019-03-12 [1] CRAN (R 3.5.2)
#> gtable 0.2.0 2016-02-26 [1] CRAN (R 3.5.1)
#> highr 0.8 2019-03-20 [1] CRAN (R 3.5.3)
#> htmltools 0.3.6 2017-04-28 [1] CRAN (R 3.5.1)
#> httr 1.4.0 2018-12-11 [1] CRAN (R 3.5.2)
#> knitr 1.22 2019-03-08 [1] CRAN (R 3.5.2)
#> labeling 0.3 2014-08-23 [1] CRAN (R 3.5.0)
#> lattice 0.20-38 2018-11-04 [2] CRAN (R 3.5.2)
#> lazyeval 0.2.2 2019-03-15 [1] CRAN (R 3.5.3)
#> magrittr 1.5 2014-11-22 [1] CRAN (R 3.5.1)
#> Matrix * 1.2-15 2018-11-01 [2] CRAN (R 3.5.2)
#> memoise 1.1.0 2017-04-21 [1] CRAN (R 3.5.1)
#> metafor * 2.0-0 2017-06-22 [1] CRAN (R 3.5.2)
#> mime 0.6 2018-10-05 [1] CRAN (R 3.5.1)
#> munsell 0.5.0 2018-06-12 [1] CRAN (R 3.5.1)
#> nlme 3.1-137 2018-04-07 [2] CRAN (R 3.5.2)
#> pillar 1.3.1 2018-12-15 [1] CRAN (R 3.5.2)
#> pkgbuild 1.0.3 2019-03-20 [1] CRAN (R 3.5.3)
#> pkgconfig 2.0.2 2018-08-16 [1] CRAN (R 3.5.1)
#> pkgload 1.0.2 2018-10-29 [1] CRAN (R 3.5.1)
#> plyr 1.8.4 2016-06-08 [1] CRAN (R 3.5.1)
#> prettyunits 1.0.2 2015-07-13 [1] CRAN (R 3.5.1)
#> processx 3.3.0 2019-03-10 [1] CRAN (R 3.5.3)
#> ps 1.3.0 2018-12-21 [1] CRAN (R 3.5.2)
#> purrr 0.3.2 2019-03-15 [1] CRAN (R 3.5.3)
#> R6 2.4.0 2019-02-14 [1] CRAN (R 3.5.2)
#> Rcpp 1.0.1 2019-03-17 [1] CRAN (R 3.5.3)
#> remotes 2.0.2 2018-10-30 [1] CRAN (R 3.5.1)
#> rlang 0.3.4 2019-04-07 [1] CRAN (R 3.5.3)
#> rmarkdown 1.12 2019-03-14 [1] CRAN (R 3.5.3)
#> rprojroot 1.3-2 2018-01-03 [1] CRAN (R 3.5.1)
#> scales 1.0.0 2018-08-09 [1] CRAN (R 3.5.1)
#> sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 3.5.1)
#> stringi 1.4.3 2019-03-12 [1] CRAN (R 3.5.2)
#> stringr 1.4.0 2019-02-10 [1] CRAN (R 3.5.1)
#> tibble 2.1.1 2019-03-16 [1] CRAN (R 3.5.3)
#> tidyselect 0.2.5 2018-10-11 [1] CRAN (R 3.5.1)
#> usethis 1.4.0 2018-08-14 [1] CRAN (R 3.5.1)
#> withr 2.1.2 2018-03-15 [1] CRAN (R 3.5.1)
#> xfun 0.5 2019-02-20 [1] CRAN (R 3.5.2)
#> xml2 1.2.0 2018-01-24 [1] CRAN (R 3.5.1)
#> yaml 2.2.0 2018-07-25 [1] CRAN (R 3.5.1)