I'm using XGBoost on a dataset of ~2.8M records of hard drive failures, where less than 200 are tagged as failures. After cleaning, there are 11 features in this dataset.
Below is my R code, as well as a link to the dataset I uploaded to my S3 bucket:
library(tidyverse)
library(caret)
library(xgboost)
library(DMwR) # SMOTE
library(Matrix)
#' Load data from S3:
ST4000DM000 <- read_csv('https://s3-us-west-2.amazonaws.com/dl.teachmehowto.trade/ST4000DM000.csv')
#' Cleaned & scaled data
dat_clean <- ST4000DM000 %>%
filter(capacity_bytes > 0) %>%
mutate_at(.vars = c("read_error_rate", "start_stop_count", "reallocated_sector", "power_on_hours", "power_cycle_count", "reported_uncorrect", "command_timeout", "high_fly_writes", "airflow_temprature", "load_cycle_count", "total_lbas_written"), # scale across vector of covariate names
funs(scale(.))) %>%
select(failure,
read_error_rate,
start_stop_count,
reallocated_sector,
power_on_hours,
power_cycle_count,
reported_uncorrect,
command_timeout,
high_fly_writes,
airflow_temprature,
load_cycle_count,
total_lbas_written)
#' Next, partition & create training/test datasets:
set.seed(42069)
idx <- createDataPartition(y = dat_clean$failure, p = 0.5, list = FALSE)
dat_train <- dat_clean[idx, ] # nrow(dat_train)
dat_test <- dat_clean[-idx, ] # nrow(dat_test)
dat_train$failure <- as.factor(dat_train$failure) # this step is required later for input into SMOTE
labels_training <- as.numeric(dat_train$failure)-1 # need the -1 because as.numeric() on factor gives 1,2
labels_test <- as.numeric(dat_test$failure)
dMtrxTrain <- xgb.DMatrix(data = model.matrix(~.+0, data = dat_train[,-1]), label = labels_training)
dMtrxTest <- xgb.DMatrix(data = model.matrix(~.+0, data = dat_test[,-1]), label = labels_test)
#' XGBoost parameters:
params.xgb <- list(booster = "gbtree",
objective = "binary:logistic",
eta = 0.3,
gamma = 0,
max_depth = 12,
min_child_weight = 1,
subsample = 1,
colsample_bytree = 1,
scale_pos_weight = 17000)
xgbcv <- xgb.cv(params = params.xgb,
data = dMtrxTrain,
nrounds = 500,
nfold = 10,
showsd = T,
stratified = T,
print_every_n = 1,
early_stopping_rounds = 20,
maximize = F)
#' Training:
model.xgb <- xgb.train(params = params.xgb,
data = dMtrxTrain,
nrounds = 100,
watchlist = list(val = dMtrxTest, train = dMtrxTrain),
print_every_n = 1,
early_stopping_rounds = 10,
maximize = F,
eval_metric = "error")
#Stopping. Best iteration:
#[44] val-error:0.000331 train-error:0.000245
#' Prediction:
xgb.pred <- predict(model.xgb, dMtrxTest)
xgb.pred <- ifelse(xgb.pred > 0.5, 1, 0)
#' Confusion Matrix
confusionMatrix(as.factor(xgb.pred), as.factor(labels_test), positive = "1")
Here's what my confusion matrix looks like:
Confusion Matrix and Statistics
Reference
Prediction 0 1
0 1410667 97
1 370 1
Accuracy : 0.9997
95% CI : (0.9996, 0.9997)
No Information Rate : 0.9999
P-Value [Acc > NIR] : 1
Kappa : 0.0042
Mcnemar's Test P-Value : <0.0000000000000002
Sensitivity : 0.0102040816
Specificity : 0.9997377815
Pos Pred Value : 0.0026954178
Neg Pred Value : 0.9999312429
Prevalence : 0.0000694476
Detection Rate : 0.0000007086
Detection Prevalence : 0.0002629089
Balanced Accuracy : 0.5049709316
'Positive' Class : 1
So, really bad. I thought to try using SMOTE to over-sample the failures:
#' SMOTE
dat_train_smote <- SMOTE(failure ~ ., data = as.data.frame(dat_train), k = 5, perc.over = 2000, perc.under = 95)
labels_smote <- as.numeric(dat_train_smote$failure)-1
dMtrxTrain_smote <- xgb.DMatrix(data = model.matrix(~.+0, data = dat_train_smote[,-1]) , label = labels_smote)
model.xgb.smote <- xgb.train(params = params.xgb,
data = dMtrxTrain_smote,
nrounds = 200,
watchlist = list(val = dMtrxTest, train = dMtrxTrain_smote),
print_every_n = 1,
early_stopping_rounds = 10,
maximize = F,
eval_metric = "error")
#' Prediction:
xgb.pred.smote <- predict(model.xgb.smote, dMtrxTest)
xgb.pred.smote <- ifelse(xgb.pred.smote > 0.5, 1, 0)
#' Confusion Matrix
confusionMatrix(as.factor(xgb.pred.smote), as.factor(labels_test), positive = "1")
Here are the results:
Confusion Matrix and Statistics
Reference
Prediction 0 1
0 1328741 49
1 82296 49
Accuracy : 0.9416
95% CI : (0.9413, 0.942)
No Information Rate : 0.9999
P-Value [Acc > NIR] : 1
Kappa : 0.0011
Mcnemar's Test P-Value : <0.0000000000000002
Sensitivity : 0.50000000
Specificity : 0.94167694
Pos Pred Value : 0.00059506
Neg Pred Value : 0.99996312
Prevalence : 0.00006945
Detection Rate : 0.00003472
Detection Prevalence : 0.05835374
Balanced Accuracy : 0.72083847
'Positive' Class : 1
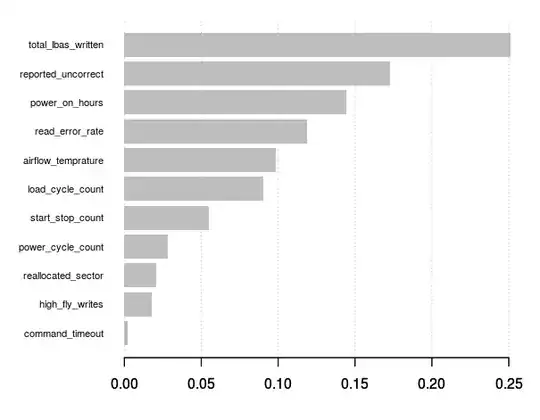
It did not improve much. In looking at a chart of variable importance however, the results seemingly "make sense" (that is, they follow my intuition about hard drives and failures):
So my question is: How can I improve this model? (if at all) What additional steps/methods should I consider?
EDIT:
Here's the ROC curve:
#' Use ROCR package to plot ROC curve & AUC
library(ROCR)
library(pROC)
xgb.perf <- performance(prediction(xgb.pred.smote, labels_test), "tpr", "fpr")
plot(xgb.perf,
avg="threshold",
colorize=TRUE,
lwd=1,
main="ROC Curve w/ Thresholds",
print.cutoffs.at=seq(0, 1, by=0.05),
text.adj=c(-0.5, 0.5),
text.cex=0.5)
grid(col="lightgray")
axis(1, at=seq(0, 1, by=0.1))
axis(2, at=seq(0, 1, by=0.1))
abline(v=c(0.1, 0.3, 0.5, 0.7, 0.9), col="lightgray", lty="dotted")
abline(h=c(0.1, 0.3, 0.5, 0.7, 0.9), col="lightgray", lty="dotted")
lines(x=c(0, 1), y=c(0, 1), col="black", lty="dotted")
roc(labels_test, xgb.pred.smote)
#Area under the curve: 0.7208