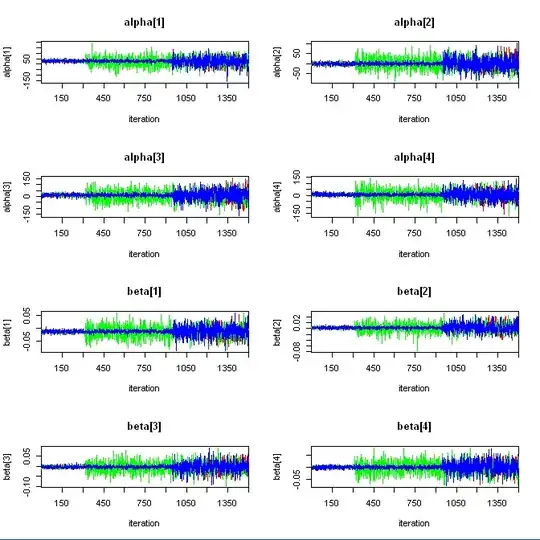
I'm trying to compute a mixed model using jags (R2jags) and I got very strange divergence. The chains started so well, very well according to the results expected (also see lmer output of the same model below). But at certain point, the chains went crazy. Just look at the traceplot for delta_tau variable - the chains start so well, but then the green chain goes crazy, then blue and finally red...
Any ideas why this happens? Can't be in initial values, because the chains started so well. Maybe the priors? Why is the system unstable?
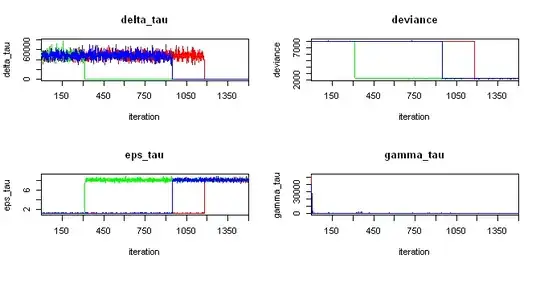
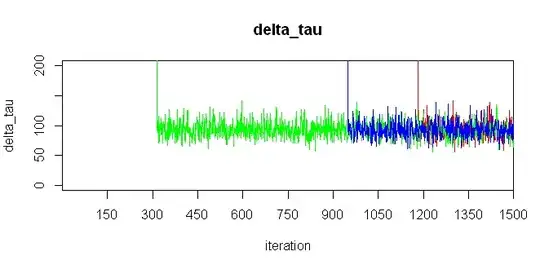
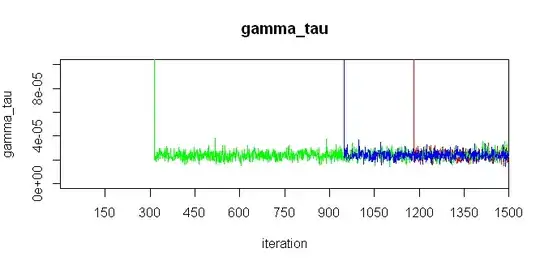
EDIT: Variables gamma_tau and delta_tau don't fall to exact zero, as you can see on these zoomed-in figures:
This is the jags model:
model {
# likelihood
for (i in 1:N) {
logInd[i] ~ dnorm(mu[i], eps_tau)
mu[i] <- alpha[crit[i]] + (beta[crit[i]] + delta[species[i]])*year[i] + gamma[species[i]] # ekviv mix1b/c podle me
}
# priors
eps_tau ~ dgamma(1.0E-3, 1.0E-3)
for (j in 1:no_crit) {
alpha[j] ~ dnorm(0, 0.0001)
beta[j] ~ dnorm(0, 0.0001)
}
for (k in 1:no_species) {
gamma[k] ~ dnorm(0, gamma_tau)
delta[k] ~ dnorm(0, delta_tau)
}
gamma_tau ~ dgamma(1.0E-3, 1.0E-3)
delta_tau ~ dgamma(1.0E-3, 1.0E-3)
}
Code used to run jags (using R2jags):
no_crit = length(levels(crit))
win.data = list(logInd = mydata$logInd, crit = (as.integer(crit)),
year = mydata$Year, species = (as.integer(mydata$Taxon)),
N = nrow(mydata), no_crit = no_crit, no_species = length(levels(mydata$Taxon))
)
inits = function () { list(
alpha = rnorm(no_crit, 0, 10000),
beta = rnorm(no_crit, 0, 10000)
)}
params = c("alpha", "beta", "eps_tau", "gamma_tau", "delta_tau")
# ni: 1000 -> .. sec
ni <- 20000
nt <- 8
nb <- 8000
nc <- 3
out <- R2jags::jags(win.data, inits, params, "model.txt",
nc, ni, nb, nt,
working.directory = paste(getwd(), "/tmp_bugs/", sep = "")
)
R2jags::traceplot(out, mfrow = c(4, 2))
Here is output from the equivalent lmer model:
> summary(lmer(logInd ~ 0 + crit_i + Year:crit_i + (1 + Year|Taxon), data = datai2))
Linear mixed model fit by REML
Formula: logInd ~ 0 + crit_i + Year:crit_i + (1 + Year | Taxon)
Data: datai2
AIC BIC logLik deviance REMLdev
8558 8630 -4267 8495 8534
Random effects:
Groups Name Variance Std.Dev. Corr
Taxon (Intercept) 1.1682e-12 1.0808e-06
Year 5.3860e-07 7.3389e-04 0.000
Residual 8.7038e-01 9.3294e-01
Number of obs: 2987, groups: Taxon, 103
Fixed effects:
Estimate Std. Error t value
crit_iA 29.0539403 8.8116915 3.297
crit_iF 0.1848404 6.0286726 0.031
crit_iU 12.3405800 10.3326242 1.194
crit_iW 5.3248537 9.7416915 0.547
crit_iA:Year -0.0122717 0.0044174 -2.778
crit_iF:Year 0.0022365 0.0030222 0.740
crit_iU:Year -0.0038701 0.0051799 -0.747
crit_iW:Year -0.0003054 0.0048836 -0.063
Correlation of Fixed Effects:
crit_A crit_F crit_U crit_W cr_A:Y cr_F:Y cr_U:Y
crit_iF 0.000
crit_iU 0.000 0.000
crit_iW 0.000 0.000 0.000
crit_iA:Yer -0.999 0.000 0.000 0.000
crit_iF:Yer 0.000 -0.999 0.000 0.000 0.000
crit_iU:Yer 0.000 0.000 -0.999 0.000 0.000 0.000
crit_iW:Yer 0.000 0.000 0.000 -0.999 0.000 0.000 0.000
Thanks in advance!