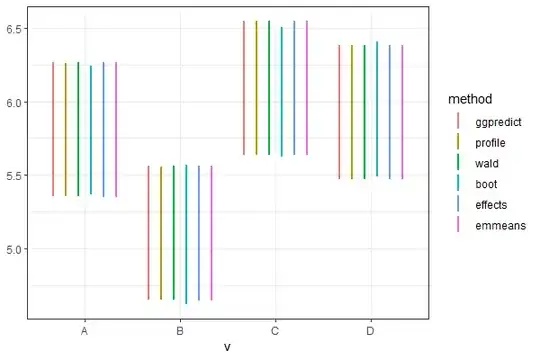
I'm using different R packages (effects, ggeffects, emmeans, lmer) to calculate confidence intervals of marginal means in a linear mixed model. My problem is that the effects package produces smaller CIs compared to other methods. Here is an example:
library(effects)
library(ggeffects)
library(emmeans)
library(lmerTest)
library(ggplot2)
ham$Age_scale <- scale(ham$Age)
# fit model without the intercept
fit <- lmer(Informed.liking ~ -1 + Product + Age_scale +
(1|Consumer/Product), data=ham)
Model summary:
Random effects:
Groups Name Variance Std.Dev.
Product:Consumer (Intercept) 3.1464 1.7738
Consumer (Intercept) 0.3908 0.6252
Residual 1.6991 1.3035
Number of obs: 648, groups: Product:Consumer, 324; Consumer, 81
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
Product1 5.809e+00 2.327e-01 3.110e+02 24.960 <2e-16 ***
Product2 5.105e+00 2.327e-01 3.110e+02 21.936 <2e-16 ***
Product3 6.093e+00 2.327e-01 3.110e+02 26.180 <2e-16 ***
Product4 5.926e+00 2.327e-01 3.110e+02 25.464 <2e-16 ***
Age_scale 9.621e-03 1.311e-01 7.900e+01 0.073 0.942
Then, I used different methods to calculate the CIs for the marginal means for Product:
term = 'Product'
## ggpredict
c0 <- as.data.frame(ggpredict(fit, terms = term))
c0 <- c0[,4:5]
## confint.merMod
c1 <- confint(fit, method='profile')
c1 <- c1[4:7,]
## confint.merMod
c2 <- confint(fit,method='Wald')
c2 <- c2[4:7,]
## confint.merMod
c3 <- confint(fit,method='boot')
c3 <- c3[4:7,]
## effect
c4 <- with(effect(term,fit),cbind(lower,upper))
## emmeans,'kenward-roger'
c5 <- with(summary(emmeans(fit,spec=term)),cbind(lower.CL,upper.CL))
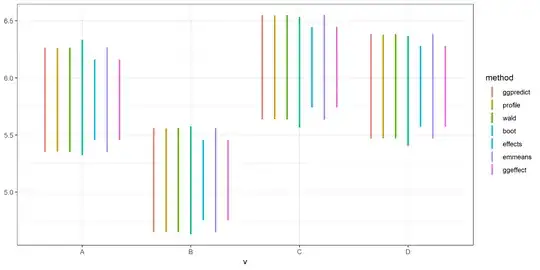
I put all the CIs together:
tmpf <- function(method,val) {
data.frame(method=method,
v=LETTERS[1:4],
setNames(as.data.frame(tail(val,4)),
c("lwr","upr")))
}
allCI <- rbind(tmpf("ggpredict",c0),
tmpf('profile',c1),
tmpf('wald',c2),
tmpf('boot',c3),
tmpf("effects",c4),
tmpf("emmeans",c5))
ggplot(allCI,aes(v,ymin=lwr,ymax=upr,colour=method))+
geom_linerange(position=position_dodge(width=0.8),size=1) + theme_bw()
The confidence intervals produced by the effects package are substantially smaller than others. I noticed someone asked a similar question before, but its answers showed that CIs produced byeffects are very similar to others. I wonder what's going on here. Did I miss something?
UPDATE: @Daniel ran the exact same code and found no deviation for the effects package. ggeffect() and ggemmeans() from version 0.8.0 of the ggeffects package also produced very similar CIs.
However, I still got smaller CIs for effects (version 4.0.1) and ggeffects (version 0.7.0), at .95 confidence level.
## effect
eff = effect(term,fit)
c4 <- with(eff,cbind(lower,upper))
## emmeans,'kenward-roger'
c5 <- with(summary(emmeans(fit,spec=term)),cbind(lower.CL,upper.CL))
## ggeffect
c6 <- ggeffect(fit, terms = term)
c6 <- c6[,4:5]
packageVersion('ggeffects')
‘0.7.0’
packageVersion('effects')
‘4.0.1’
eff$confidence.level
0.95
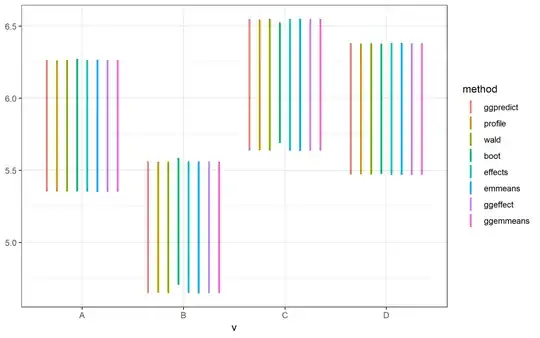
After upgrading to the latest versions, however, all methods produced similar CIs.
packageVersion('ggeffects')
‘0.8.0’
packageVersion('effects')
‘4.1.0’
## effect
eff = effect(term,fit)
c4 <- with(eff,cbind(lower,upper))
## emmeans,'kenward-roger'
c5 <- with(summary(emmeans(fit,spec=term)),cbind(lower.CL,upper.CL))
## ggeffect
c6 <- ggeffect(fit, terms = term)
c6 <- c6[,4:5]
## ggemmeans (only the 0.8.0 version supports this)
c7 <- ggemmeans(fit, terms = term)
c7 <- c7[,4:5]