I have data with 6 predictor variables and a response variable (default ). When I do a logistic regression using probit estimation, I get a result different from SAS.
(the link to my data https://drive.google.com/open?id=1TZvEsOpuzLQ_BvGBR26n5M-yzlp2yUWH)
equation <- as.formula(default ~ dlagR3+loglaginf6+dloglagImp6+loglagRR9+dlagFX6+laglogRGDP6 )
model <- glm(equation , family = binomial('probit'), data=setDF(data))
But R is giving the following error:
glm.fit: fitted probabilities numerically 0 or 1 occurred
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -5.13523 1.55705 -3.298 0.000974 ***
dlagR3 0.04719 0.03696 1.277 0.201711
loglaginf6 -0.39395 0.05637 -6.989 2.76e-12 ***
dloglagImp6 0.13151 0.02981 4.412 1.02e-05 ***
loglagRR9 6.78246 0.09228 73.497 < 2e-16 ***
dlagFX6 0.02099 0.01835 1.144 0.252535
laglogRGDP6 -1.52335 0.32653 -4.665 3.08e-06 ***
But when I use SAS or Eviews, this error is not thrown and they both agree on the coefficients.
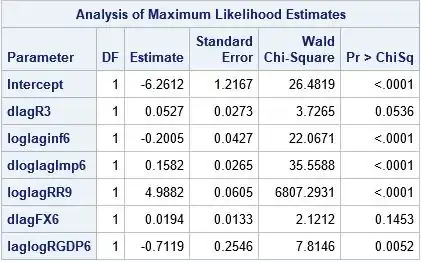
So I tried implementing a lot of supposed solutions as given in a lot of forums but no matter what I do, R is giving the same coefficients. The results given by Eviews and SAS is the accurate one.
Note: SAS gives the probability of 0 (not 1) so I have created a new response variable default2 which is opposite of default
proc logistic data=data;
model default2 = dlagR3 loglaginf6 dloglagImp6 loglagRR9 dlagFX6 laglogRGDP6
/link=probit;
run;