I have the following survival data: cases with a continuous variable risk_factor wich higher values having a higher HR (1.026, CI: [1.017-1.034], p = 2.03e-09) for survival (after running regression Cox model). I want to verify the log-linearity of the continuous variable risk_factor for predicting the survival. How could I do it? What would be the explanation and the code. Does it make sense to do it? Can it reinforce the information given by the Cox regression model?
library(broom)
library(tidyr)
timespan_censored = c(round(runif(450, 0, 4500), digit = 0),
round(runif(150, 0, 1200), digit = 0))
risk_factor = c(runif(450, min=10, max=80),
runif(150, min=20, max=100))
status = c(rep(0, 450), rep(1, 150))
timespan_censored = c(round(runif(450, 0, 4500), digit = 0),
round(runif(150, 0, 1200), digit = 0))
df_try = data.frame(status, timespan_censored, risk_factor)
cox_mod = coxph(Surv(timespan_censored, status) ~ risk_factor, data = df_try) %>% summary()
cox_mod
this is the output:
Call:
coxph(formula = Surv(timespan_censored, status) ~ risk_factor,
data = df_try)
n= 600, number of events= 150
coef exp(coef) se(coef) z Pr(>|z|)
risk_factor 0.025294 1.025617 0.004219 5.995 2.03e-09 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
exp(coef) exp(-coef) lower .95 upper .95
risk_factor 1.026 0.975 1.017 1.034
Concordance= 0.629 (se = 0.024 )
Rsquare= 0.062 (max possible= 0.953 )
Likelihood ratio test= 38.28 on 1 df, p=6.133e-10
Wald test = 35.94 on 1 df, p=2.032e-09
Score (logrank) test = 37.31 on 1 df, p=1.007e-09
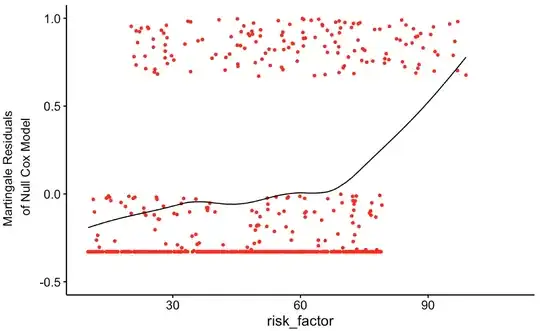
ggcoxfunctional(Surv(timespan_censored, status) ~ risk_factor, data = df_try, xlim = c(10,110), ylim = c(-0.5,1.0))
ggcoxfunctional(Surv(timespan_censored, status) ~ log(risk_factor), data = df_try, xlim = c(2,5), ylim = c(-0.35,1.0))
ggcoxfunctional(Surv(timespan_censored, status) ~ sqrt(risk_factor), data = df_try, xlim = c(3,10), ylim = c(-0.35,1.0))
library(rms)
cox_mod_spline = coxph(Surv(timespan_censored,status)~ rcs(risk_factor,6))
cox_mod_spline
res = residuals(cox_mod_spline, type = "martingale")
plot(df_try$risk_factor, res)
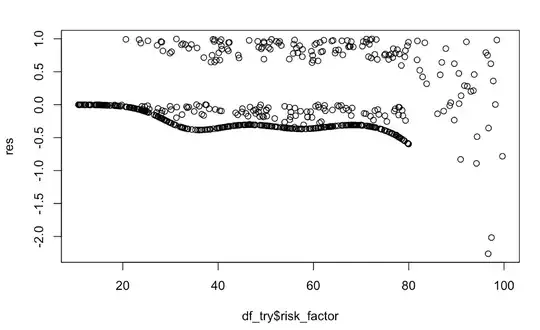
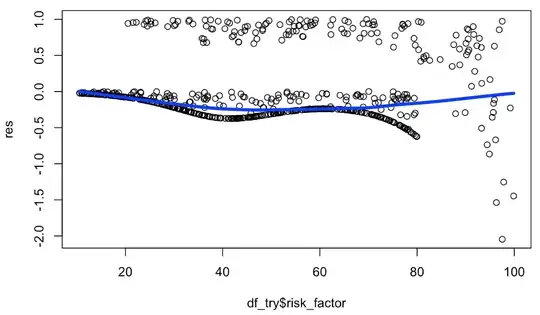
cox_mod_spline = coxph(Surv(timespan_censored,status)~ rcs(risk_factor,4))
cox_mod_spline
res = residuals(cox_mod_spline, type = "martingale")
plot(df_try$risk_factor, res)
lines(loess.smooth(df_try$risk_factor, res), lwd = 4, col="blue")