I want to simulate data to test a logistic regression. The main problem I have is the error term when I generate the binomial response. Although, I know (see other posts here and here) we are modelling the mean so there is no error in the link function, I'm getting a warning that my GLM fits perfectly the data:
Warning message:
glm.fit: fitted probabilities numerically 0 or 1 occurred
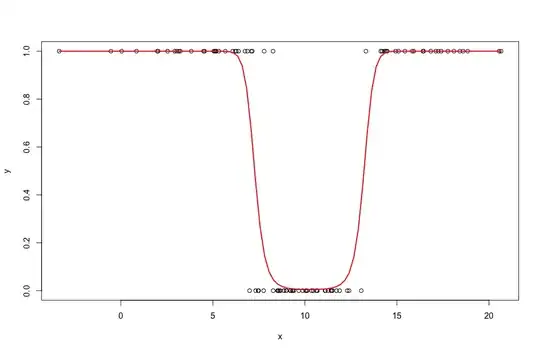
But I want to generate data that is more realistic (e.g. when looking at survival of individuals in biology, we do not expect a perfect separation 000011111 but more something like 001011101). Here is what I have so far:
set.seed(1345)
n.1 =100
mean.1 = 10
sd.1 = 5
sd.erro = 0.5
x.1 = rnorm(n.1, mean.1, sd.1)
x = c(x.1)
n = length(x)
mn = mean(x)
sg = sd(x)
z = 1 - 1*x + 1*(x-mean(x))^2 + rnorm(length(x),
mean = 0,
sd = sd.erro)
pr = 1/(1+exp(-z))
y = rbinom(length(x),1,pr)
df = data.frame(y = y,
x = x,
x.2 = x^2)
hist(x)
hist(residuals(lm(z~x)), breaks = 30)
par(mfrow = c(2,1))
plot(z~x)
abline(v = c(mn,mn+sg,mn-sg), lty = 2)
# As you can see there is a perfect separation in the reponse:
plot(y~x)
# Generate the warning
model = glm(y~ x + x.2,
data = df,
family="binomial")
newx = data.frame(x = seq(min(x),
max(x),
length.out = n))
newx$x.2 = newx$x^2
newx$y = predict(model,
newx,
type="response")
logi = function(x) {exp(x)/(1+exp(x))}
newx$y.logi = logi(newx$y)
lines(x = newx$x,
y = newx$y,
col = "red",
lwd=2,
ylim = c(0,1))
So the idea here is where should I add an error term that will generate a more variable response. Is it only the sd.errothat I need to modify?