You can test significance, of model parameters, with the help of estimated confidence intervals for which the lme4 package has the confint.merMod function.
bootstrapping (see for instance Confidence Interval from bootstrap)
> confint(m, method="boot", nsim=500, oldNames= FALSE)
Computing bootstrap confidence intervals ...
2.5 % 97.5 %
sd_(Intercept)|participant_id 0.32764600 0.64763277
cor_conditionexperimental.(Intercept)|participant_id -1.00000000 1.00000000
sd_conditionexperimental|participant_id 0.02249989 0.46871800
sigma 0.97933979 1.08314696
(Intercept) -0.29669088 0.06169473
conditionexperimental 0.26539992 0.60940435
likelihood profile (see for instance What is the relationship between profile likelihood and confidence intervals?)
> confint(m, method="profile", oldNames= FALSE)
Computing profile confidence intervals ...
2.5 % 97.5 %
sd_(Intercept)|participant_id 0.3490878 0.66714551
cor_conditionexperimental.(Intercept)|participant_id -1.0000000 1.00000000
sd_conditionexperimental|participant_id 0.0000000 0.49076950
sigma 0.9759407 1.08217870
(Intercept) -0.2999380 0.07194055
conditionexperimental 0.2707319 0.60727448
There is also a method 'Wald' but this is applied to fixed effects only.
There also exist some kind of anova (likelihood ratio) type of expression in the package lmerTest which is named ranova. But I can not seem to make sense out of this. The distribution of the differences in logLikelihood, when the null hypothesis (zero variance for the random effect) is true is not chi-square distributed (possibly when number of participants and trials is high the likelihood ratio test might make sense).
Variance in specific groups
To obtain results for variance in specific groups you could reparameterize
# different model with alternative parameterization (and also correlation taken out)
fml1 <- "~ condition + (0 + control + experimental || participant_id) "
Where we added two columns to the data-frame (this is only needed if you wish to evaluate non-correlated 'control' and 'experimental' the function (0 + condition || participant_id) would not lead to the evaluation of the different factors in condition as non-correlated)
#adding extra columns for control and experimental
d <- cbind(d,as.numeric(d$condition=='control'))
d <- cbind(d,1-as.numeric(d$condition=='control'))
names(d)[c(4,5)] <- c("control","experimental")
Now lmer will give variance for the different groups
> m <- lmer(paste("sim_1 ", fml1), data=d)
> m
Linear mixed model fit by REML ['lmerModLmerTest']
Formula: paste("sim_1 ", fml1)
Data: d
REML criterion at convergence: 2408.186
Random effects:
Groups Name Std.Dev.
participant_id control 0.4963
participant_id.1 experimental 0.4554
Residual 1.0268
Number of obs: 800, groups: participant_id, 40
Fixed Effects:
(Intercept) conditionexperimental
-0.114 0.439
And you can apply the profile methods to these. For instance now confint gives confidence intervals for the control and exerimental variance.
> confint(m, method="profile", oldNames= FALSE)
Computing profile confidence intervals ...
2.5 % 97.5 %
sd_control|participant_id 0.3490873 0.66714568
sd_experimental|participant_id 0.3106425 0.61975534
sigma 0.9759407 1.08217872
(Intercept) -0.2999382 0.07194076
conditionexperimental 0.1865125 0.69149396
Simplicity
You could use the likelihood function to get more advanced comparisons, but there are many ways to make approximations along the road (e.g. you could do a conservative anova/lrt-test, but is that what you want?).
At this point it makes me wonder what is actually the point of this (not so common) comparison between variances. I wonder whether it starts to become too sophisticated. Why the difference between variances instead of the ratio between variances (which relates to the classical F-distribution)? Why not just report confidence intervals? We need to take a step back, and clarify the data and the story it is supposed to tell, before going into advanced pathways that may be superfluous and loose touch with the statistical matter and the statistical considerations that are actually the main topic.
I wonder whether one should do much more than simply stating the confidence intervals (which may actually tell much more than a hypothesis test. a hypothesis test gives a yes no answer but no information about the actual spread of the population. given enough data you can make any slight difference to be reported as a significant difference). To go more deeply into the matter (for whatever purpose), requires, I believe, a more specific (narrowly defined) research question in order to guide the mathematical machinery to make the proper simplifications (even when an exact calculation might be feasible or when it could be approximated by simulations/bootstrapping, even then in in some settings it still requires some appropriate interpretation). Compare with Fisher's exact test to solve a (particular) question (about contingency tables) exactly, but which may not be the right question.
Simple example
To provide an example of the simplicity that is possible I show below a comparison (by simulations) with a simple assessment of the difference between the two group variances based on an F-test done by comparing variances in the individual mean responses and done by comparing the mixed model derived variances.
For the F-test we simply compare the variance of the values (means) of the individuals in the two groups. Those means are for condition $j$ distributed as:
$$\hat{Y}_{i,j} \sim N(\mu_j, \sigma_j^2 + \frac{\sigma_{\epsilon}^2}{10})$$
if the measurement error variance $\sigma_\epsilon$ is equal for all individuals and conditions, and if the variance for the two conditions $\sigma_{j}$ (with $j = \lbrace 1,2 \rbrace$) is equal then the ratio for the variance for the 40 means in the condition 1 and the variance for the 40 means in the condition 2 is distributed according to the F-distribution with degrees of freedom 39 and 39 for numerator and denominator.
You can see this in the simulation of the below graph where aside for the F-score based on sample means an F-score is calculated based on the predicted variances (or sums of squared error) from the model.
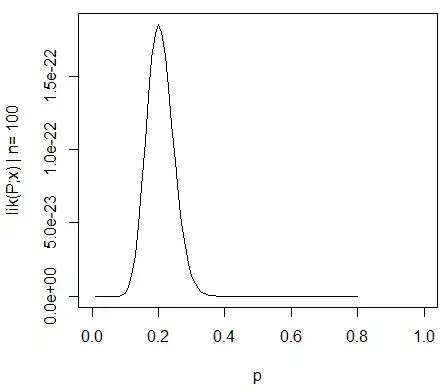
The image is modeled with 10 000 repetitions using $\sigma_{j=1} = \sigma_{j=2} = 0.5$ and $\sigma_\epsilon=1$.
You can see that there is some difference. This difference may be due to fact that the mixed effects linear model is obtaining the sums of squared error (for the random effect) in a different way. And these squared error terms are not (anymore) well expressed as a simple Chi-squared distribution, but still closely related and they can be approximated.
Aside from the (small) difference when the null-hypothesis is true, more interesting is the case when the null hypothesis is not true. Especially the condition when $\sigma_{j=1} \neq \sigma_{j=2}$. The distribution of the means $\hat{Y}_{i,j}$ are not only dependent on those $\sigma_j$ but also on the measurement error $\sigma_\epsilon$. In the case of the mixed effects model this latter error is 'filtered out', and it is expected that the F-score based on the random effects model variances has a higher power.
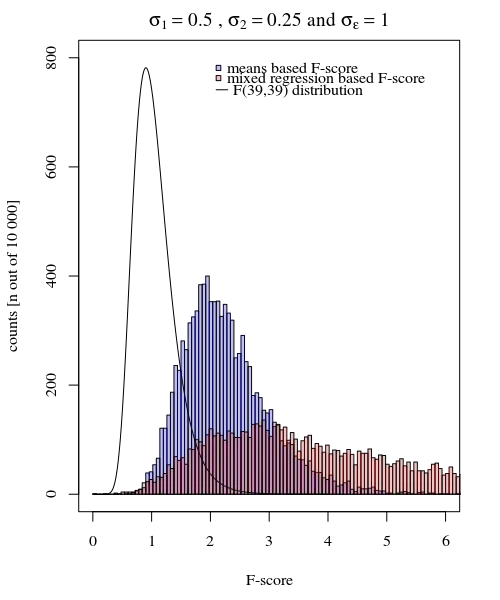
The image is modeled with 10 000 repetitions using $\sigma_{j=1} = 0.5$, $\sigma_{j=2} = 0.25$ and $\sigma_\epsilon=1$.
So the model based on the means is very exact. But it is less powerful. This shows that the correct strategy depends on what you want/need.
In the example above when you set the right tail boundaries at 2.1 and 3.1 you get approximately 1% of the population in the case of equal variance (resp 103 and 104 of the 10 000 cases) but in the case of unequal variance these boundaries differ a lot (giving 5334 and 6716 of the cases)
code:
set.seed(23432)
# different model with alternative parameterization (and also correlation taken out)
fml1 <- "~ condition + (0 + control + experimental || participant_id) "
fml <- "~ condition + (condition | participant_id)"
n <- 10000
theta_m <- matrix(rep(0,n*2),n)
theta_f <- matrix(rep(0,n*2),n)
# initial data frame later changed into d by adding a sixth sim_1 column
ds <- expand.grid(participant_id=1:40, trial_num=1:10)
ds <- rbind(cbind(ds, condition="control"), cbind(ds, condition="experimental"))
#adding extra columns for control and experimental
ds <- cbind(ds,as.numeric(ds$condition=='control'))
ds <- cbind(ds,1-as.numeric(ds$condition=='control'))
names(ds)[c(4,5)] <- c("control","experimental")
# defining variances for the population of individual means
stdevs <- c(0.5,0.5) # c(control,experimental)
pb <- txtProgressBar(title = "progress bar", min = 0,
max = n, style=3)
for (i in 1:n) {
indv_means <- c(rep(0,40)+rnorm(40,0,stdevs[1]),rep(0.5,40)+rnorm(40,0,stdevs[2]))
fill <- indv_means[d[,1]+d[,5]*40]+rnorm(80*10,0,sqrt(1)) #using a different way to make the data because the simulate is not creating independent data in the two groups
#fill <- suppressMessages(simulate(formula(fml),
# newparams=list(beta=c(0, .5),
# theta=c(.5, 0, 0),
# sigma=1),
# family=gaussian,
# newdata=ds))
d <- cbind(ds, fill)
names(d)[6] <- c("sim_1")
m <- lmer(paste("sim_1 ", fml1), data=d)
m
theta_m[i,] <- m@theta^2
imeans <- aggregate(d[, 6], list(d[,c(1)],d[,c(3)]), mean)
theta_f[i,1] <- var(imeans[c(1:40),3])
theta_f[i,2] <- var(imeans[c(41:80),3])
setTxtProgressBar(pb, i)
}
close(pb)
p1 <- hist(theta_f[,1]/theta_f[,2], breaks = seq(0,6,0.06))
fr <- theta_m[,1]/theta_m[,2]
fr <- fr[which(fr<30)]
p2 <- hist(fr, breaks = seq(0,30,0.06))
plot(-100,-100, xlim=c(0,6), ylim=c(0,800),
xlab="F-score", ylab = "counts [n out of 10 000]")
plot( p1, col=rgb(0,0,1,1/4), xlim=c(0,6), ylim=c(0,800), add=T) # means based F-score
plot( p2, col=rgb(1,0,0,1/4), xlim=c(0,6), ylim=c(0,800), add=T) # model based F-score
fr <- seq(0, 4, 0.01)
lines(fr,df(fr,39,39)*n*0.06,col=1)
legend(2, 800, c("means based F-score","mixed regression based F-score"),
fill=c(rgb(0,0,1,1/4),rgb(1,0,0,1/4)),box.col =NA, bg = NA)
legend(2, 760, c("F(39,39) distribution"),
lty=c(1),box.col = NA,bg = NA)
title(expression(paste(sigma[1]==0.5, " , ", sigma[2]==0.5, " and ", sigma[epsilon]==1)))