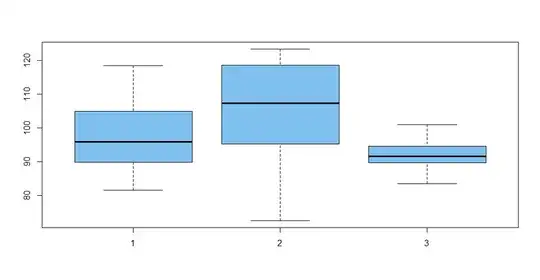
There are five groups and I want to test for difference in means between the five groups assuming the variance are not constant between the groups.
-
3Look at `oneway.test()` as a possible solution. – ekstroem Jul 13 '18 at 21:21
-
4How about Welch's anova? – Sal Mangiafico Jul 14 '18 at 03:02
-
Also, see @gung 's [answer here](https://stats.stackexchange.com/questions/91872/alternatives-to-one-way-anova-for-heteroskedastic-data/91881#91881) for different approaches including white-adjusted anova, weighted least squares, and sandwich estimators. – Sal Mangiafico Jul 14 '18 at 15:47
-
Thank you everyone for helping me with these type of questions. – Hello Jul 14 '18 at 17:11
2 Answers
One of several possible methods is to use oneway.test in R.
Here is an example with three groups, each with ten observations:
x1 = rnorm(10, 100, 10); x2 = rnorm(10, 95, 15); x3 = rnorm(10, 90, 5)
x = c(x1, x2, x3); group = rep(1:3, each=10)
boxplot(x ~ group)
You can see that my fake data were generated with different standard deviations in each of the three groups. The boxplots show this heteroscedasticity. A Bartlett test confirms significance (P-value 0.004).
sd(x1); sd(x2); sd(x3)
[1] 10.88923
[1] 16.35099
[1] 4.656118
bartlett.test(x, group)
Bartlett test of homogeneity of variances
data: x and group
Bartlett's K-squared = 11.103, df = 2, p-value = 0.003881
The oneway.test procedure allows for different variances in somewhat the
same way as does the Welch 2-sample t test. It indicates that not all
group population means are equal (P-value < 5%). Notice that the denominator DF $\approx 15;$ a standard ANOVA assuming equal variances would have denominator DF $= 27.$ [I believe this is the 'Welch ANOVA' suggested by @SalMangiafico.]
mean(x1); mean(x2); mean(x3)
[1] 98.00458
[1] 105.3806
[1] 92.0077
oneway.test(x ~ group)
One-way analysis of means (not assuming equal variances)
data: x and group
F = 3.818, num df = 2.000, denom df = 14.536, p-value = 0.04649
You could use Welch 2-sample t tests to explore paired comparisons, perhaps with a Bonferroni family error rate.
Reference: This Q & A mentions a variety of alternative methods.
- 47,896
- 2
- 28
- 76
-
3For what it's worth, I would not recommend the Kruskal-Wallis (KW) test. To look at a difference in medians for the KW-test, you would have to assume that the only difference between the groups is the medians. All groups have to be identically shaped, this assumption includes having similar variances. Only normality is irrelevant. – Heteroskedastic Jim Jul 14 '18 at 14:22
-
1It's not that Kruskal-Wallis isn't applicable or desirable in this situation. It's just that it isn't (usually) a test of medians. K-W, as a test of stochastic equality, is often very useful and meaningful... Also, the question asked specifically about means. – Sal Mangiafico Jul 14 '18 at 15:42
-
1It is quite an interesting issue. KW is a test of dominance. If you have unequal variances, then you may find that one group dominates the other regardless of the mean differences. So it is when one extends the claim to a median difference that homogeneity of variance matters. So the original page did not make this claim about medians based on my quick check. – Heteroskedastic Jim Jul 14 '18 at 15:43
-
I see that oneway.test requires a normal distribution assumption. How can one proceed if the normality assumption is not met? – Hello Jul 14 '18 at 17:12
-
1Probably not. // Depending on circumstances, I might try one of the alternative methods in the link at the end of my Answer. I'd have to see the data and understand the situation. // I might try a permutation test. Perhaps using the 'Welch ANOVA' F-statistic as metric, but using its permutation distribution, rather than an F-distribution, to get the P-value. (Perhaps see [Eudey et al (2010)](http://ww2.amstat.org/publications/jse/v18n1/eudey.pdf) for an elementary introduction the permutation tests; 2-sample tests are discussed in Sect. 4. A similar approach would work for a one-way ANOVA.) – – BruceET Jul 14 '18 at 17:45
-
I just posted a new question describing my situation. Thank you very much for willing me to help. – Hello Jul 14 '18 at 17:58
I would like to recommend generalized least squares as an option. Same way you can create a model for the mean of the data given your predictors, you can also create a model for variance. The gls() function in the nlme package permits us to do this.
I will demo how below:
set.seed(1984)
n <- 25
# Create heteroskedastic data, balanced design
dat <- data.frame(
y = c(
rnorm(n, 1, 1), rnorm(n, 0, 1), rnorm(n, .5, 1),
rnorm(n, 0, 3), rnorm(n, -.5, 4)),
groups = sort(rep(LETTERS[1:5], n))
)
boxplot(y ~ groups, dat)
# Assuming all cases have identical variances
summary(aov(y ~ groups, dat))
Df Sum Sq Mean Sq F value Pr(>F)
groups 4 69.0 17.255 3.475 0.0101 *
Residuals 120 595.9 4.966
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
# Modeling the error variances
library(nlme)
# This is your standard linear model
coef(summary(fit.gls.baseline <- gls(y ~ groups, dat))))
Value Std.Error t-value p-value
(Intercept) 1.1784414 0.4457005 2.644021 0.0092875812
groupsB -0.9748277 0.6303157 -1.546571 0.1246000669
groupsC -0.8980932 0.6303157 -1.424831 0.1568017302
groupsD -1.2903790 0.6303157 -2.047195 0.0428223361
groupsE -2.3076197 0.6303157 -3.661054 0.0003747902
sigma(fit.gls.baseline) # get residual standard deviation
[1] 2.228502
# Next we fit a heteroskedastic model, hidden some output
# varIdent says the variances are identical for cases that have the same value of group
summary(fit.gls <- gls(
y ~ groups, dat, weights = varIdent(form = ~ 1 | groups)))
Generalized least squares fit by REML
Model: y ~ groups
Data: dat
AIC BIC logLik
479.5354 507.4103 -229.7677
Variance function:
Structure: Different standard deviations per stratum
Formula: ~1 | groups
Parameter estimates:
A B C D E
1.0000000 0.9741804 0.8698579 3.0062743 3.7867899
Coefficients:
Value Std.Error t-value p-value
(Intercept) 1.1784414 0.1951410 6.038923 0.0000
groupsB -0.9748277 0.2724316 -3.578248 0.0005
groupsC -0.8980932 0.2586375 -3.472401 0.0007
groupsD -1.2903790 0.6182517 -2.087142 0.0390
groupsE -2.3076197 0.7642898 -3.019299 0.0031
Residual standard error: 0.975705
A few notes about this model. You will find the residual standard deviation (error) is much less for the heteroskedastic model when compared to the same estimate for the earlier model. Now take a look at the Variance function section, and you will see group A had a standard deviation of 1. These values are relative standard deviations. So the $.976$ reported for the model is the residual SD of group A. $.974 \times .976$ is the value for group B. Groups D $(3 \times .976)$ and E $(3.79 \times .976)$ have much larger residual standard deviations.
Also, let's obtain the heteroskedastic version of the F-test:
# marginal for type III sum of squares, only makes a difference for the
# test of the grouping variable if we have more than one grouping variable
anova(fit.gls, type = "marginal")
Denom. DF: 120
numDF F-value p-value
(Intercept) 1 36.46859 <.0001
groups 4 5.51477 4e-04
Sufficient evidence to suggest that not all groups share the same mean, $F(4, 120)=5.51, p < .001$.
Now to compare the heteroskedastic model to the standard model, note that the coefficients are about the same, however, the standard errors are much smaller for the heteroskedastic model, except for groups D and E. So our p-values are also smaller. A global approach to model comparison would be:
anova(fit.gls, fit.gls.baseline) # Model fit improved
Model df AIC BIC logLik Test L.Ratio p-value
fit.gls 1 10 479.5354 507.4103 -229.7677
fit.gls.baseline 2 6 560.9588 577.6837 -274.4794 1 vs 2 89.42343 <.0001
These results suggest the heteroskedastic model was better, lower AIC, BIC and statistically significant likelihood ratio test.
Now, we do not have access to the data generation process in reality, but we can always plot the boxplt above. Assuming we want to opt for a simpler model, we can assume groups A, B and C have the same variances and groups D and E are different. We can then try:
# Create a variable that classifies the groups according to variance
dat$var.groups <- (dat$groups %in% c("D", "E")) + 0
# then run:
summary(fit.gls.parsim <- gls(
y ~ groups, dat, weights = varIdent(form = ~ 1 | var.groups)))
Generalized least squares fit by REML
Model: y ~ groups
Data: dat
AIC BIC logLik
475.3162 494.8287 -230.6581
Variance function:
Structure: Different standard deviations per stratum
Formula: ~1 | var.groups
Parameter estimates:
0 1
1.000000 3.600023
Coefficients:
Value Std.Error t-value p-value
(Intercept) 1.1784414 0.1853218 6.358893 0.0000
groupsB -0.9748277 0.2620846 -3.719516 0.0003
groupsC -0.8980932 0.2620846 -3.426730 0.0008
groupsD -1.2903790 0.6924235 -1.863569 0.0648
groupsE -2.3076197 0.6924235 -3.332671 0.0011
# A model comparison between our previous best and this new model
anova(fit.gls, fit.gls.parsim)
Model df AIC BIC logLik Test L.Ratio p-value
fit.gls 1 10 479.5354 507.4103 -229.7677
fit.gls.parsim 2 7 475.3162 494.8287 -230.6581 1 vs 2 1.780859 0.6191
The likelihood ratio test cannot distinguish between this simpler model that has three fewer parameters and our earlier heteroskedastic model. Also AIC and BIC lean towards this model. So we may opt for this model going forward since we do not know the truth. Theory may also suggest certain things about the differences in group variances. One can see how the particular issue of heteroskedasticity can be of substantive interest in itself.
F-test again would be:
anova(fit.gls.parsim, type = "marginal")
Denom. DF: 120
numDF F-value p-value
(Intercept) 1 40.43552 <.0001
groups 4 6.04201 2e-04
Now, the next thing may be contrasts. I do not care much for this in my work, so I do the simplest things here. There are probably better ways to deal with contrasts, see the emmeans package documentation for more options. I just use defaults.
library(emmeans)
pairs(emmeans(fit.gls.parsim, "groups"))
contrast estimate SE df t.ratio p.value
A - B 0.9748277 0.2620846 120 3.720 0.0028
A - C 0.8980932 0.2620846 120 3.427 0.0073
A - D 1.2903790 0.6924235 120 1.864 0.3427
A - E 2.3076197 0.6924235 120 3.333 0.0099
B - C -0.0767345 0.2620846 120 -0.293 0.9984
B - D 0.3155513 0.6924235 120 0.456 0.9910
B - E 1.3327920 0.6924235 120 1.925 0.3099
C - D 0.3922858 0.6924235 120 0.567 0.9796
C - E 1.4095265 0.6924235 120 2.036 0.2555
D - E 1.0172407 0.9435106 120 1.078 0.8174
P value adjustment: tukey method for comparing a family of 5 estimates
These contrasts use information from the heteroskedastic model as the results would be different if you used the fit.gls.baseline model.
I hope the demonstration above shows how you can account for heteroskedasticity using generalized least squares when performing analysis of variance. Compared to many other approaches, this approach does not simply treat heteroskedasticity as a nuisance. One can also include control variables as in ANCOVA.
As an aside, if one also suspects that there is also heteroskedasticity by the control variables in ANCOVA, then the glmmTMB() function in the glmmTMB package can handle this situation. You simply specify a regression equation for the variance using the dispformula = argument.
- 4,567
- 1
- 10
- 32
-
My scenario is that I have 40 different brands and I am recording 10 variables for that 40 brands Each brand was observed 6 times. My first step was to find sample standard deviation by each brand, so that I can group by equal variance. I used PCA to do that. Then I performed cluster analysis to find optimal number of clusters. Now let's say I have 5 clusters that means I have 5 different groups. Would I be able to run gls model as you mentioned based on 5 clusters that I have now? – Hello Jul 14 '18 at 17:15
-
1This scenario is very different from what you described in your question. I am not sure I understand all you write here. However, creating clusters to use as predictors in a new model definitely creates bias in your regression estimates, if only because of measurement error in the clusters you have created. If you are intent on running the ANOVA, then sure, you can use `gls()` if heteroskedasticity is a concern. But if I were reviewing your analysis, I would not be satisfied with the series of steps you have laid out. I would suggest you ask a new question laying out all you are doing. – Heteroskedastic Jim Jul 14 '18 at 17:35
-
1I just posted a new question. Thank you very much for willing me to help. – Hello Jul 14 '18 at 17:57