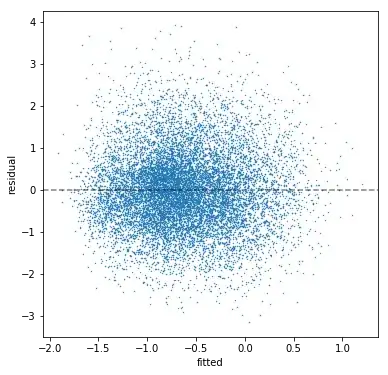
I'm working in Python with statsmodels. I estimate a multiple regression model (n=10763; 12 predictors; r^2=0.216; all coefficients have signs pointing the correct direction and are significant). Then I check my residuals. The residuals show no discernible pattern, so there appears to be negligible heteroskedasticity:
The residuals have a Jarque-Bera test statistic of 338.7 with a p-value of 2e-75. The skew is 0.317 and the kurtosis is 3.543. We can see this in a histogram of the residuals:
And in the residuals' q-q plot:
I have two questions, relating to satisfying the OLS assumption that the residuals have a normal distribution to get meaningful p-values.
The first is how to interpret if your residuals are "close enough" to normally distributed. Presumably, a sufficiently large sample size will always fail the J-B test and other related tests -- i.e., producing a p-value below .05 so we can reject the null hypothesis that the residuals are normally distributed. But looking at the histogram and the q-q plot, how close to "perfect" do we expect them to be? They seem to show the thin tails and slight right skew. Given real-world data, what is "good enough"?
Second question: what sort of model re-specifications or corrections would one typically explore to correct for residuals distributed with high kurtosis?