The agricolae::HSD.test function does exactly that, but you will need to let it know that you are interested in an interaction term. Here is an example with a Stata dataset:
library(foreign)
yield <- read.dta("http://www.stata-press.com/data/r12/yield.dta")
tx <- with(yield, interaction(fertilizer, irrigation))
amod <- aov(yield ~ tx, data=yield)
library(agricolae)
HSD.test(amod, "tx", group=TRUE)
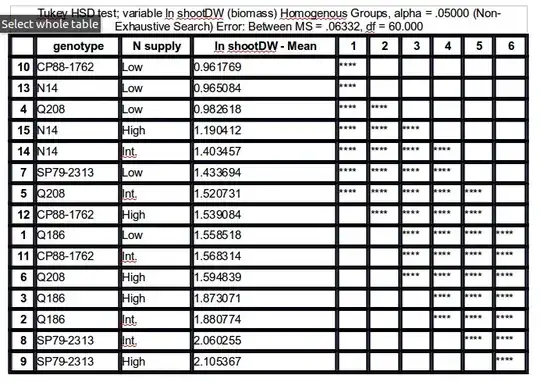
This gives the results shown below:
Groups, Treatments and means
a 2.1 51.17547
ab 4.1 50.7529
abc 3.1 47.36229
bcd 1.1 45.81229
cd 5.1 44.55313
de 4.0 41.81757
ef 2.0 38.79482
ef 1.0 36.91257
f 3.0 36.34383
f 5.0 35.69507
They match what we would obtain with the following commands:
. webuse yield
. regress yield fertilizer##irrigation
. pwcompare fertilizer#irrigation, group mcompare(tukey)
-------------------------------------------------------
| Tukey
| Margin Std. Err. Groups
----------------------+--------------------------------
fertilizer#irrigation |
1 0 | 36.91257 1.116571 AB
1 1 | 45.81229 1.116571 CDE
2 0 | 38.79482 1.116571 AB
2 1 | 51.17547 1.116571 F
3 0 | 36.34383 1.116571 A
3 1 | 47.36229 1.116571 DEF
4 0 | 41.81757 1.116571 BC
4 1 | 50.7529 1.116571 EF
5 0 | 35.69507 1.116571 A
5 1 | 44.55313 1.116571 CD
-------------------------------------------------------
Note: Margins sharing a letter in the group label are
not significantly different at the 5% level.
The multcomp package also offers symbolic visualization ('compact letter displays', see Algorithms for Compact Letter Displays: Comparison and Evaluation for more details) of significant pairwise comparisons, although it does not present them in a tabular format. However, it has a plotting method which allows to conveniently display results using boxplots. Presentation order can be altered as well (option decreasing=), and it has lot more options for multiple comparisons. There is also the multcompView package which extends those functionalities.
Here is the same example analyzed with glht:
library(multcomp)
tuk <- glht(amod, linfct = mcp(tx = "Tukey"))
summary(tuk) # standard display
tuk.cld <- cld(tuk) # letter-based display
opar <- par(mai=c(1,1,1.5,1))
plot(tuk.cld)
par(opar)
Treatment sharing the same letter are not significantly different, at the chosen level (default, 5%).
Incidentally, there is a new project, currently hosted on R-Forge, which looks promising: factorplot. It includes line and letter-based displays, as well as a matrix overview (via a level plot) of all pairwise comparisons. A working paper can be found here: factorplot: Improving Presentation of Simple Contrasts in GLMs