If you had a straight-up linear model, this would just be a question of scaling the residuals. For a binomial model, it's a bit harder. I think your best best will likely be to do rejection sampling. Simulate your model many times and pick out the one that yields a p value closest to what you want. (Simply store the RNG seeds so you can recreate it.)
Note that with $n=1000$ observations and rather strong effects, your p values may well be far lower than the 0.01 you are aiming for, so you might want to reduce either $n$ or the effect size.
n.obs <- 20
n.sims <- 1e4
p.values <- rep(NA,n.sims)
for ( ii in 1:n.sims ) {
set.seed(ii)
x1 = rnorm(n.obs) # some continuous variables
x2 = rnorm(n.obs)
z = 1 + 2*x1 + 3*x2 # linear combination with a bias
pr = 1/(1+exp(-z)) # pass through an inv-logit function
y = rbinom(n.obs,1,pr) # bernoulli response variable
#now feed it to glm:
df = data.frame(y=y,x1=x1,x2=x2)
model <- glm( y~x1+x2,data=df,family="binomial")
p.values[ii] <- summary(model)$coefficients[2,4]
}
hist(p.values,breaks=seq(0,1,by=0.01))
which.min(abs(p.values-0.01))
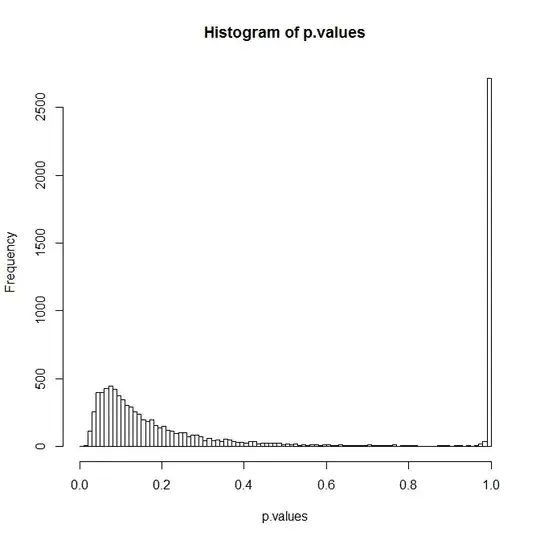
The big peak at 1 probably comes from those simulations where you got all zeros or all ones. You can now reset the RNG using the output from the last line and resimulate your model:
set.seed(which.min(abs(p.values-0.01)))
x1 = rnorm(n.obs) # some continuous variables
x2 = rnorm(n.obs)
z = 1 + 2*x1 + 3*x2 # linear combination with a bias
pr = 1/(1+exp(-z)) # pass through an inv-logit function
y = rbinom(n.obs,1,pr) # bernoulli response variable
#now feed it to glm:
df = data.frame(y=y,x1=x1,x2=x2)
summary(glm( y~x1+x2,data=df,family="binomial"))
The result:
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.0701 0.8382 1.277 0.2017
x1 1.7253 0.6888 2.505 0.0122 *
x2 1.1760 0.8392 1.401 0.1611
