It is best to get the data into a normalized form where smoking and heart attack are separate, parallel columns and other columns provide the identifying (key) fields:
country year smoking heart
1 Congo 1988 1200 900
2 Congo 1984 1146 400
3 Congo 2010 675 550
4 Nigeria 1988 1100 950
5 Nigeria 1984 786 568
6 Nigeria 2010 765 590
7 Zimbabwe 1988 1098 1098
8 Zimbabwe 1984 897 769
9 Zimbabwe 2010 900 865
In R they ought to be in a data frame:
data <- data.frame(country=c(rep("Congo", 3), rep("Nigeria", 3), rep("Zimbabwe", 3)),
year=rep(c(1988, 1984, 2010), 3),
smoking=c(1200, 1146, 675, 1100, 786, 765, 1098, 897, 900),
heart=c(900,400, 550, 950, 568, 590, 1098, 769, 865))
With this format, the solution is a one-liner:
by(data, data$country, function(x) cor(x$smoking, x$heart))
The output gives correlations of 0.315, 0.994, and 0.962 for Congo, Nigeria, and Zimbabwe, respectively.
However, correlations by themselves can be misleading. It is far better to look at the data:
plot(data$smoking, data$heart, col=data$country, pch=19,
main="Heart attack and smoking, 1984-2010", xlab="Smoking", ylab="Heart attacks")
points(data$smoking, data$heart, cex=1.25)
legend(675, 1075, levels(data$country), pch=19, col=1:length(levels(data$country)))
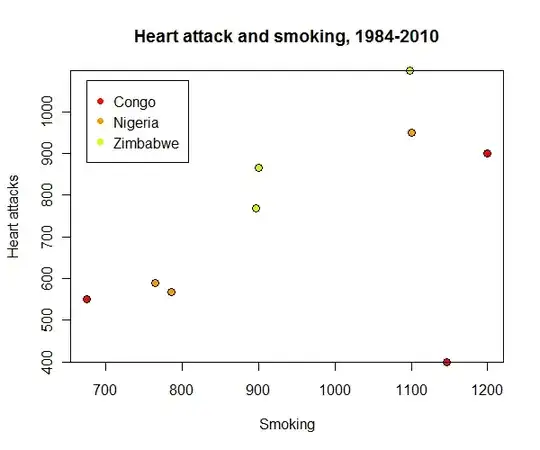
An outlying value for Congo visible in the lower right at (1146, 400) is responsible for its low correlation coefficient: the correlation is a poor description of the smoking-heart attack relation for this country's data.
