Using R, I have built a linear model from the fluorescence produced by a set of genetic sequences under certain experimental conditions.
The final linear model provides a number of different metrics about the model, such as RMSE, R-Squared, the estimates of the predictors' coefficients, standard errors, t-statistics, and p-values. What I would like to do is take the predictors' coefficients and standard errors and use them to predict the fluorescence of a new set of sequences whose actual fluorescence values have yet to be experimentally determined.
I know that one can acquire the model's coefficients and standard errors by running the command summary(model)$coefficients to get an output similar to:
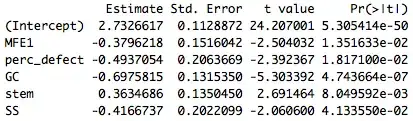
However, I am particularly confused as to how I can get a standard error of the whole model that I can use to make the confidence interval of the predicted outcome. I know that one can use confint(model, level=0.95) in order to get the upper and lower limits of the predictors, but I don't know whether it is accurate to take these numbers and use them to determine the confidence interval of the predicted outcome.
To illustrate, I would calculate the final outcome by taking the in silica calculations of new sequences and plugging them into the final model:
predicted_outcome= -0.3807*MFE1 + -0.4937*perc_defect + -0.6976*GC + 0.3635*stem + -0.4167*SS + 2.733
Then, I thought that in order to calculate the upper and lower limits of the confidence intervals, I would use values calculated by confint(model, level=0.95) to determine the predicted outcome's confidence interval.
predicted_lower_limit= -0.6768*MFE1 + -0.89817*perc_defect + -0.9553*GC + 0.0987*stem + -0.8129*SS + 2.511
predicted_upper_limit= -0.0825*MFE1 + -0.0892*perc_defect + -0.4398*GC + 0.6282*stem + -0.0203*SS + 2.954
Because I am working between building the model in R and implementing on a data frame in Python, I want to be able to calculate the predicted values and their corresponding confidence intervals on my own. Furthermore, I am not sure as to whether this approach is the proper way of understanding the confidence interval of a predicted outcome using a model. Can anybody confirm or correct this approach?