Here is a sample of 20 rows from some data I'm working with (everything below is consistent with the full dataset):
lat cond id trial
1388 0 9627850 Pleasing|
2102 1 9654467 Disgust|
747 0 9644328 Happy|
2112 0 9638350 wmwf53.jpg|
579 1 9639422 Tragic|
2175 1 9628964 Despise|
800 1 9653958 wmwf53.jpg|
1276 0 9640461 wmwf52.jpg|
1602 1 9636965 Fabulous|
1140 0 9642953 wmbf4.jpg|
1070 1 9633495 bmbf3.jpg|
1000 1 9654108 Horrible|
473 1 9640532 Smiling|
476 1 9617722 Glorious|
891 1 9636287 bmbf31.jpg|
908 0 9635345 wmwf53.jpg|
592 0 9631025 bmbf40.jpg|
704 1 9642356 bmbf40.jpg|
756 1 9645445 wmwf39.jpg|
854 1 9646231 Cheerful|
I'm trying to model latency ("lat") as a function of condition ("cond") while taking into account random effects of person ("id") and stimulus type ("trial").I notice that in the full dataset, that latency is positively skewed:
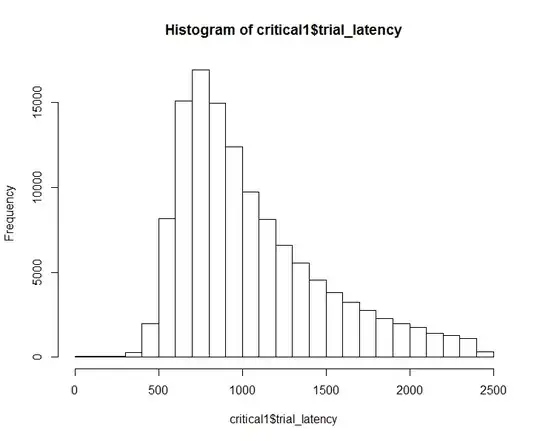 Even in the sample data posted here with 20 observations, it seems like a GLM might be a better way to go (Model 1):
Even in the sample data posted here with 20 observations, it seems like a GLM might be a better way to go (Model 1):
Call:
lm(formula = lat ~ cond, data = v)
Residuals:
Min 1Q Median 3Q Max
-574.1 -354.6 -164.6 137.9 1137.9
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1166.1 207.5 5.621 2.47e-05 ***
cond -129.1 257.3 -0.502 0.622
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 548.9 on 18 degrees of freedom
Multiple R-squared: 0.01378, Adjusted R-squared: -0.04101
F-statistic: 0.2516 on 1 and 18 DF, p-value: 0.622
Compared with gamma distribution (Model 2):
Call:
glm(formula = lat ~ cond, family = Gamma, data = v)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.6945 -0.3766 -0.1681 0.1134 0.8445
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.0008575 0.0001664 5.153 6.67e-05 ***
cond 0.0001067 0.0002157 0.495 0.627
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for Gamma family taken to be 0.2636089)
Null deviance: 4.2975 on 19 degrees of freedom
Residual deviance: 4.2341 on 18 degrees of freedom
AIC: 307.59
Number of Fisher Scoring iterations: 5
Model comparison BIC (Model1, Model2):
df BIC
Model1 3 315.9530
Model2 3 310.5728
Similarly, in the full dataset, the GLM is a much better fit.
Then, I next tried to add a random effect of trial. Lmer worked fine:
Linear mixed model fit by REML ['lmerMod']
Formula: lat ~ cond + (1 | trial)
Data: v
REML criterion at convergence: 282.7
Scaled residuals:
Min 1Q Median 3Q Max
-1.0460 -0.6460 -0.2998 0.2512 2.0732
Random effects:
Groups Name Variance Std.Dev.
trial (Intercept) 0 0.0
Residual 301274 548.9
Number of obs: 20, groups: trial, 17
Fixed effects:
Estimate Std. Error t value
(Intercept) 1166.1 207.5 5.621
cond -129.1 257.3 -0.502
Correlation of Fixed Effects:
(Intr)
cond -0.806
Glmer did not:
> mod2 <- glmer(lat ~ cond + (1|trial),
+ data=v,family=Gamma)
Warning messages:
1: In checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0250853 (tol = 0.001,
component 1)
2: In checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model is nearly unidentifiable: very large eigenvalue
- Rescale variables?
I know that this is just a small sample of my data, but the error messages are the same for the entire dataset. Thus, although I have a distribution that should be better fit with GLMER, why am I getting error messages?