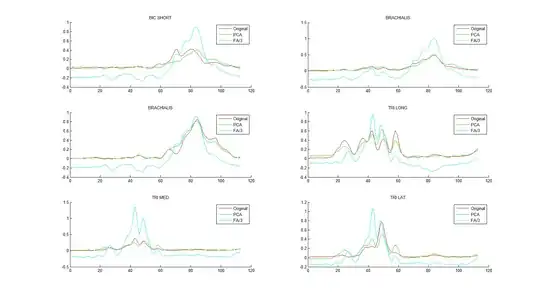
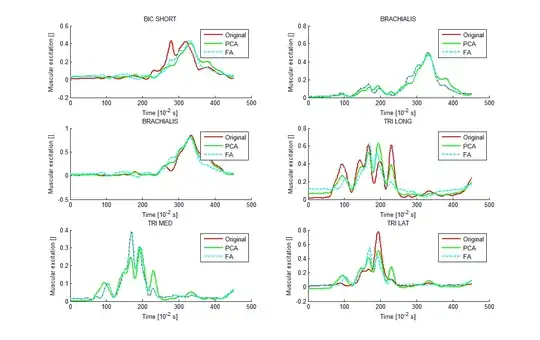
I saw this interesting topic: How to reverse PCA and reconstruct original variables from several principal components? and a nice answer with a very useful example of Iris data in Matlab. I would like to do the same using factor analysis instead of PCA. I tried to make it with 'factoran' of Matlab with the help of @ttnphns and @amoeba but I don't obtain a good correlation between my reconstructed data and the original ones.
input_data (*data are EMG measurement from 6 arm muscles in order to identify synergies)
PCA method:
X = input_data;
mu = mean(X);
[eigenvectors, scores] = pca(X);
nComp = 2;
Xpca = scores(:,1:nComp) * eigenvectors(:,1:nComp)';
Xpca = bsxfun(@plus, Xpca, mu);
I obtain good correlation between them.
FA method:
X = input_data;
mu = mean(X);
[LoadingsPM,specVarPM,rotationPM,stats, scores] = ...
factoran(X,2,'rotate','promax');
Xfa = scores*LoadingsPM';
Xfa = bsxfun(@plus, Xfa, mu);
But in this case the correlations are bad. I don't know if I forget something? (I divided per 3 the FA reconstruction in order to see better the 3 curves).
@ttnphns note: word "reverse" in the title should be taken in the technical sense of computing variables as they are returned by the computed factors (their scores), - not in the theoretical sense (in which FA model is nothing but predicting variables by factors, so that there is no a "reverse" direction). In PCA, this prediction/direction indeed could be called "reverse" in a theoretical sense, too.