Question
Is it correct to test the difference of a measurement between groups using generalized least squares?
Example
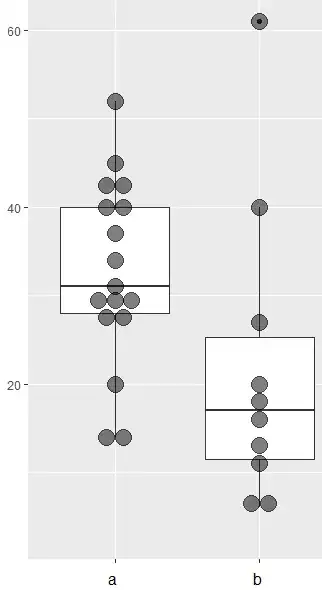
My data looks like this:
I perform a gls in R, but I`m not sure if it is a correct method. Usually I dont have categorical variables and I just applied these functions because I always do.
lsmeans(gls(outcome ~ group, data = mydata, pairwise ~ group, adjust="tukey")
$lsmeans
group lsmean SE df lower.CL upper.CL
a 32.64706 3.219347 25 26.01669 39.27743
b 21.90000 4.197515 25 13.25506 30.54494
Confidence level used: 0.95
$contrasts
contrast estimate SE df t.ratio p.value
a - b 10.74706 5.289927 25 2.032 0.0530
Here group is a factor and outcome is numeric. If this is a correct approach, what would this test be called?
Alternative
If I perform a t test, which might be more correct after thinking about it, the P value is doubled compared to the method above. Is that due to the method, or due to the fact that one reports "a minus b" and the other "a compared to b" ?
t.test(outcome ~ group, mydata)
Welch Two Sample t-test
data: outcome by group
t = 1.8015, df = 13.182, p-value = 0.09454
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-2.123109 23.617227
sample estimates:
mean in group a mean in group b
32.64706 21.90000