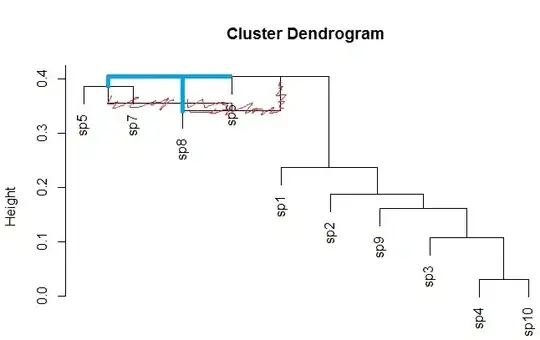
I'm attempting to perform hierarchical agglomerative cluster analysis in R.
However, when I use particular clustering methods, I get reversals (upward branching) in the resulting tree, which violates the ultrametric property.
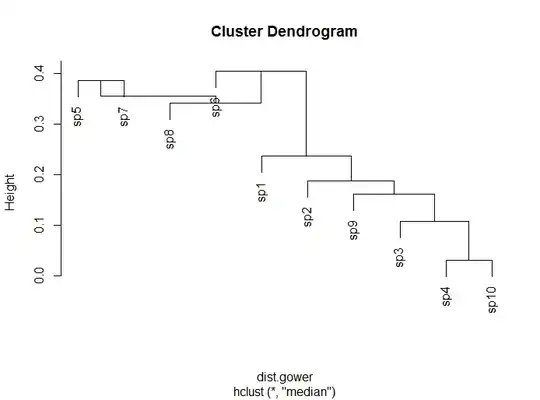
The two methods are: UPGMC and WPGMC (methods="median" and "centroid" in hclust). Legendre & Legendre in their Numerical Ecology book suggest some reasons why this may occur (Section 8.6). However, they provide no solutions to rectify the issue and convert the trees to ultrametric.
I'm curious: is this an unavoidable consequence of the data and the clustering method, or is there a way that I can produce a tree that satisfies the ultrametric property using these two methods?
Here is an example data set and R code to play with:
#Generate data frame with mixed continuous and categorical trait data for 10 species
set.seed(91)
(df=data.frame(trait1=runif(10,0,10),trait2=runif(10,0,10),
trait3=sample(letters[1:3],10,replace=T),row.names=paste("sp",1:10,sep="")))
#Generate Gower dissimilarity matrix from trait data
library(cluster)
(dist.gower=daisy(df,metric="gower"))
#Create a vector of clustering methods
tree.methods=c("ward","single","complete","average","mcquitty","median","centroid")
#Build the trees using each method
trees=lapply(tree.methods,function(i) hclust(dist.gower,method=i))
#Plot the trees
par(mfrow=c(4,2))
for(i in 1:length(trees)) {plot(trees[[i]])}
#The last two trees have reversals...cannot be converted to ultrametric!