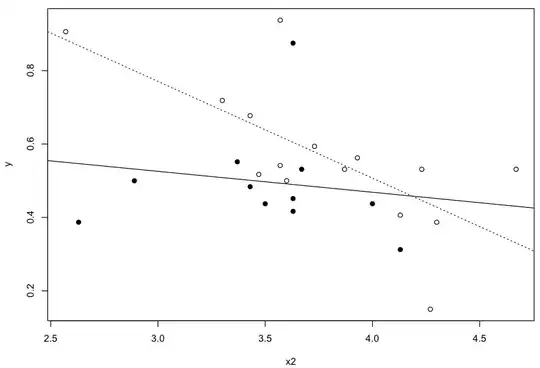
Consider the following example:
df <- data.frame(y=c(0.4375, 0.4167, 0.5313, 0.4516, 0.5417, 0.5172,
0.1500, 0.5161, 0.5313, 0.5000, 0.4839, 0.3871,
0.3125, 0.5313, 0.4063, 0.5517, 0.3871, 0.7188,
0.7188, 0.5484, 0.9375, 0.5938, 0.4375, 0.8750,
0.9063, 0.6774, 0.5625, 0.5000, 0.5313),
x1=c("B", "B", "B", "B", "A", "A", "A", "B", "A",
"A", "B", "A", "B", "A", "A", "B", "B", "B",
"A", "B", "A", "A", "B", "B", "A", "A", "A",
"B", "A"),
x2=c(4.00, 3.63, 3.67, 3.63, 3.57, 3.47, 4.27,
2.17, 3.87, 3.60, 3.43, 4.30, 4.13, 4.67,
4.13, 3.37, 2.63, 2.33, 3.30, 2.33, 3.57, 3.73,
3.50, 3.63, 2.57, 3.43, 3.93, 2.89, 4.23))
plot(y ~ x2, data=subset(df, x1=="A"))
abline(lm(y ~ x2, data=subset(df, x1=="A")), lty=3)
points(y ~ x2, data=subset(df, x1=="B"), pch=19)
abline(lm(y ~ x2, data=subset(df, x1=="B")))
l <- lm(y ~ x1 * x2, data=df)
summary(l)
So the interaction of x1 * x2 is significantly different from zero:
Call:
lm(formula = y ~ x1 * x2, data = df)
Residuals:
Min 1Q Median 3Q Max
-0.28588 -0.06650 -0.01718 0.03695 0.38534
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.56204 0.28445 5.491 1.05e-05 ***
x1B -0.86511 0.34959 -2.475 0.02047 *
x2 -0.26374 0.07469 -3.531 0.00163 **
x1B:x2 0.20664 0.09683 2.134 0.04283 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.1436 on 25 degrees of freedom
Multiple R-squared: 0.3648, Adjusted R-squared: 0.2886
F-statistic: 4.786 on 3 and 25 DF, p-value: 0.009052
Now I would like to see the correlation coefficients for each level of predictor x1:
r1 <- sqrt(summary(lm(y ~ x2, data=subset(df, x1=="A")))$r.squared)
r2 <- sqrt(summary(lm(y ~ x2, data=subset(df, x1=="B")))$r.squared)
I'm using Fisher's r-to-z transformation to compare the coefficients and would expect them to be significantly different as well, reflecting the interaction in my previous linear model that showed different slopes for x2 depending on the level of x1:
z1 <- (1/2) * (log(1+r1) - log(1-r1))
n1 <- nrow(subset(df, x1=="A"))
n2 <- nrow(subset(df, x1=="B"))
z2 <- (1/2) * (log(1+r2) - log(1-r2))
z <- (z1 - z2) / (sqrt((1/(n1 - 3)) + (1/(n2 - 3))))
However, this is not what I found; the z = 1.41, which is not significantly different from zero (P = 0.16):
pval <- 2 * pnorm(-abs(z))
I'm wondering why this is the case. Shouldn't the correlation coefficients be significantly different?