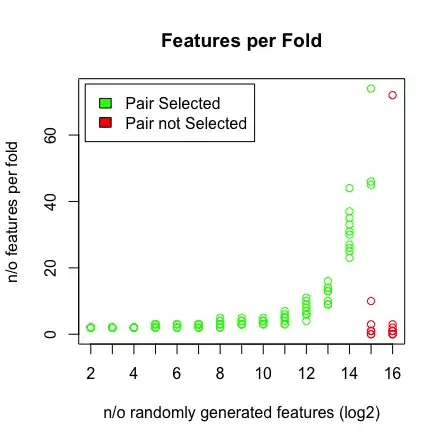
I'm running a small experiment with LASSO regression in R to test if it is able to find a perfect predictor pair. The pair is defined like this: f1 + f2 = outcome
The outcome here is a predetermined vector called 'age'. F1 and f2 are created by taking half of the age vector and setting the rest of the values to 0, for example: age = [1,2,3,4,5,6], f1 = [1,2,3,0,0,0] and f2 = [0,0,0,4,5,6]. I combine this predictor pair with an increasing amount of randomly created variables by sampling from a normal distribution N(1,1).
What I see is when I hit 2^16 variables, LASSO is not finding my pair anymore. See the results below.
Why is this happening? You can reproduce the results with the script below. I've noticed that when I pick a different age vector, e.g: [1:193] then LASSO does find the pair at high dimensionality (>2^16).
The Script:
## Setup ##
library(glmnet)
library(doParallel)
library(caret)
mae <- function(errors){MAE <- mean(abs(errors));return(MAE)}
seed = 1
n_start <- 2 #start at 2^n features
n_end <- 16 #finish with 2^n features
cl <- makeCluster(3)
registerDoParallel(cores=cl)
#storage of data
features <- list()
coefs <- list()
L <- list()
P <- list()
C <- list()
RSS <- list()
## MAIN ##
for (j in n_start:n_end){
set.seed(seed)
age <- c(55,31,49,47,68,69,53,42,58,67,60,58,32,52,63,31,51,53,37,48,31,58,36,42,61,49,51,45,61,57,52,60,62,41,28,45,39,47,70,33,37,38,32,24,66,54,59,63,53,42,25,56,70,67,44,33,50,55,60,50,29,51,49,69,70,36,53,56,32,43,39,43,20,62,46,65,62,65,43,40,64,61,54,68,55,37,59,54,54,26,68,51,45,34,52,57,51,66,22,64,47,45,31,47,38,31,37,58,66,66,54,56,27,40,59,63,64,27,57,32,63,32,67,38,45,53,38,50,46,59,29,41,33,40,33,69,42,55,36,44,33,61,43,46,67,47,69,65,56,34,68,20,64,41,20,65,52,60,39,50,67,49,65,52,56,48,57,38,48,48,62,48,70,55,66,58,42,62,60,69,37,50,44,61,28,64,36,68,57,59,63,46,36)
beta2 <- as.data.frame(cbind(age,replicate(2^(j),rnorm(length(age),1,1))));colnames(beta2)[1] <-'age'
f1 <- c(age[1:96],rep(0,97))
f2 <- c(rep(0,96),age[97:193])
beta2 <- as.data.frame(cbind(beta2,f1,f2))
#storage variables
L[[j]] <- vector()
P[[j]] <- vector()
C[[j]] <- list()
RSS[[j]] <- vector()
#### DCV LASSO ####
set.seed(seed) #make folds same over 10 iterations
for (i in 1:10){
print(paste(j,i))
index <- createFolds(age,k=10)
t.train <- beta2[-index[[i]],];row.names(t.train) <- NULL
t.test <- beta2[index[[i]],];row.names(t.test) <- NULL
L[[j]][i] <- cv.glmnet(x=as.matrix(t.train[,-1]),y=as.matrix(t.train[,1]),parallel = T,alpha=1)$lambda.min #,lambda=seq(0,10,0.1)
model <- glmnet(x=as.matrix(t.train[,-1]),y=as.matrix(t.train[,1]),lambda=L[[j]][i],alpha=1)
C[[j]][[i]] <- coef(model)[,1][coef(model)[,1] != 0]
pred <- predict(model,as.matrix(t.test[,-1]))
RSS[[j]][i] <- sum((pred - t.test$age)^2)
P[[j]][i] <- mae(t.test$age - pred)
gc()
}
}
##############
## PLOTTING ##
##############
#calculate plots features
beta_sum = unlist(lapply(unlist(C,recursive = F),function(x){sum(abs(x[-1]))}))
penalty = unlist(L) * beta_sum
RSS = unlist(RSS)
pair_coefs <- unlist(lapply(unlist(C,recursive = F),function(x){
if('f1' %in% names(x)){f1 = x['f1']}else{f1=0;names(f1)='f1'}
if('f2' %in% names(x)){f2 = x['f2']}else{f2=0;names(f2)='f2'}
return(c(f1,f2))}));pair_coefs <- split(pair_coefs,c('f1','f2'))
inout <- lapply(unlist(C,recursive = F),function(x){c('f1','f2') %in% names(x)})
colors <- unlist(lapply(inout,function(x){if (x[1]*x[2]){'green'}else{'red'}}))
featlength <- unlist(lapply(unlist(C,recursive = F),function(x){length(x)-1}))
#diagnostics
plot(rep(n_start:n_end,each=10),pair_coefs$f1,col='red',xaxt = "n",xlab='n/o randomly generated features (log2)',main='Pair Coefficients',ylim=c(0,1),ylab='pair coefficients');axis(1, at=n_start:n_end);points(rep(n_start:n_end,each=10),pair_coefs$f2,col='blue');axis(1, at=n_start:n_end, labels=(n_start:n_end));legend('bottomleft',fill=c('red','blue'),legend = c('f1','f2'),inset=.02)
plot(rep(n_start:n_end,each=10),RSS+penalty,col=colors,xaxt = "n",xlab='n/o randomly generated features (log2)',main='RSS+penalty');axis(1, at=n_start:n_end, labels=(n_start:n_end));legend('topleft',fill=c('green','red'),legend = c('Pair Selected','Pair not Selected'),inset=.02)
plot(rep(n_start:n_end,each=10),penalty,col=colors,xaxt = "n",xlab='n/o randomly generated features (log2)',main='Penalty');axis(1, at=n_start:n_end, labels=(n_start:n_end));legend('topleft',fill=c('green','red'),legend = c('Pair Selected','Pair not Selected'),inset=.02)
plot(rep(n_start:n_end,each=10),RSS,col=colors,xaxt = "n",xlab='n/o randomly generated features (log2)',main='RSS');axis(1, at=n_start:n_end, labels=(n_start:n_end));legend('topleft',fill=c('green','red'),legend = c('Pair Selected','Pair not Selected'),inset=.02)
plot(rep(n_start:n_end,each=10),unlist(L),col=colors,xaxt = "n",xlab='n/o randomly generated features (log2)',main='Lambdas',ylab=expression(paste(lambda)));axis(1, at=n_start:n_end, labels=(n_start:n_end));legend('topleft',fill=c('green','red'),legend = c('Pair Selected','Pair not Selected'),inset=.02)
plot(rep(n_start:n_end,each=10),featlength,ylab='n/o features per fold',col=colors,xaxt = "n",xlab='n/o randomly generated features (log2)',main='Features per Fold');axis(1, at=n_start:n_end, labels=(n_start:n_end));legend('topleft',fill=c('green','red'),legend = c('Pair Selected','Pair not Selected'),inset=.02)
plot(penalty,RSS,col=colors,main='Penalty vs. RSS')