More specifically, I have two predictor (age with three levels and sex with two levels) treated as factors and one response (salary). I want to test if age, sex and their interaction are significant related to salary. After conducting type III two-way anova (unbalanced observations),
age <-as.factor(c(2,3,2,4,2,3,4,3,4,4,3,3,2,2,2,4,4,3,3,3,2,2,2,3,4,4,2))
sex <- as.factor(c(1,0,0,0,0,1,1,1,0,1,0,1,1,0,1,1,0,1,1,0,0,1,0,0,1,0,0))
salary<-c(12,17,10,24,13,19,22,20,31,32,23,16,8,14,9,39,37,11,14,22,11,7,11,16,12,49,15)
mod<- lm(salary~age*sex)
library(car)
Anova(mod, type = "III")
plot(mod)
I found that the graph of "residuals vs fitted value" has heteroscedasticity with respect to its variance. So I want to use gls() function in R to make the variance more constant (homoscedasticity). What I did by coding in R was:
mod_gls = gls(salary ~ age*sex)
summary(mod_gls)
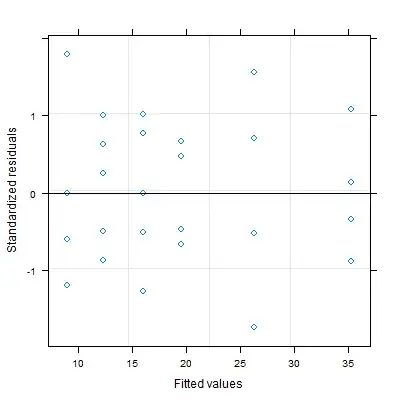
plot(mod_gls)
However, this plot gave me a plot called "standardized residual vs fitted values but still have almost same pattern as the first plot.
So I was wondering if I did gls() wrong and what is the appropriate way?