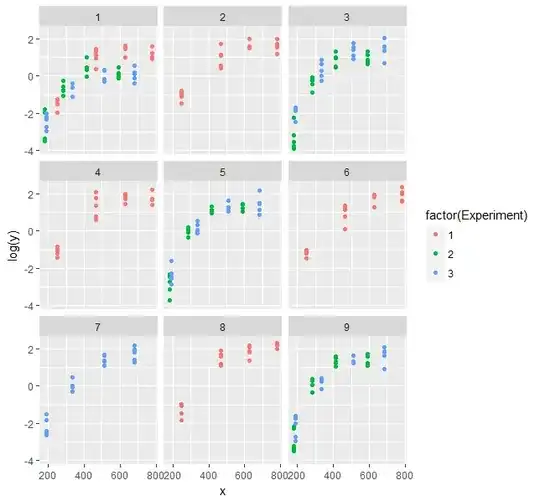
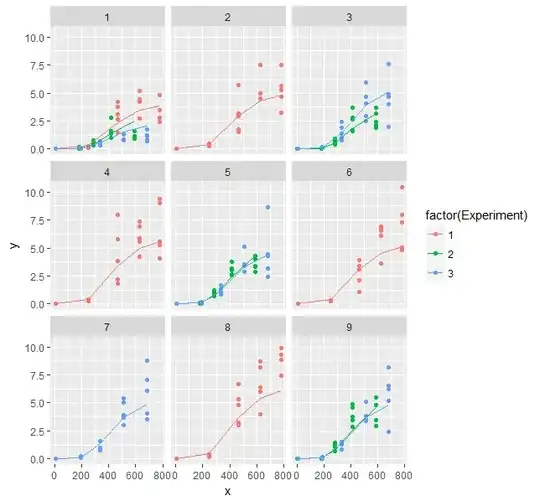
I would appreciate any help. I am trying to fit a nonlinear mixed model to some experiments. I have adopted a Gompertz function (y=Asym*exp(-b2*b3^x)), and the data used is 3 years of experiments (the doses were not always the same and I removed some outliers before any analysis). Moreover, since the measurements were destructive, it is not a repeated measure case. I am fitting a nested model in R (nlme package, tried with lme4 as well). The data are here. The final model adopted by now is:
model <- nlme(y ~ SSgompertz(x,Asym,b2,b3),
data = Data,
fixed = Asym + b2 + b3 ~ 1,
random = Asym ~ 1|Experiment/Treatment,
start = c(Asym = 5, b2 = 30, b3 = 0.8),
verbose = T, na.action = na.omit, method="ML",
control = nlmeControl(maxIter=100000, opt="nlminb", pnlsTol=0.01))
In relation to diagnosis plots:
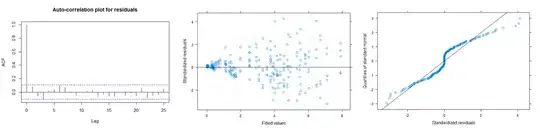
- There does not seem to be a normality problem.
- There isn't a problem with autocorrelation. However, if I can include an autocorrelation structure between groups, I think the model can be improved.
- There is increasing variability. So including a heterogeneous variance for the error term will improve the model and will help to deal with the heteroscedasticity.
The summary of the model gives a high negative correlation between b2 and b3 parameters. I am not sure how bad is this.
So my questions is: How can I correctly specify a correlation structure? I have been trying different ways and methods for the correlation function without success. On the other hand, the inclusion of weights and more random effects did not improve the model. I was planning to include a covariate but this option has not been implemented for nested models in nlme. So, in addition to specifying a correlation structure, how could I improve the model's results?