I'm wondering what the best way to generate prediction intervals from a linear model when there is known heteroscedastic AND the heteroscedasticity is predictable.
Consider the following toy example:
#generate sample data
sample_df <- data.frame(
response = c(
rnorm(50,100,20),
rnorm(50,20,5)
),
group = c(
rep("A", 50),
rep("B", 50)
)
)
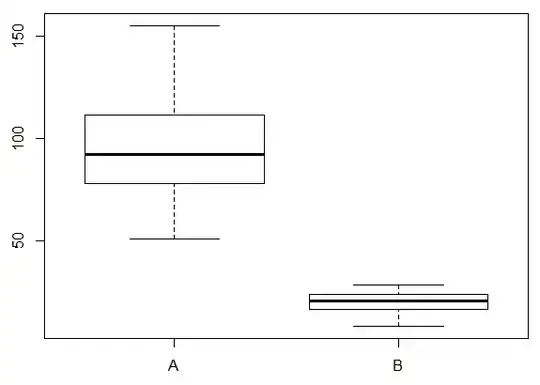
#Boxplot
boxplot(response~group, data=sample_df)
An ordinary linear model will give fine spot estimates/predictions for groups A and B, but confidence and prediction intervals will be inappropriate.
Having a look at similar questions on this topic, I've ascertained that gls() from the nlme package can be used to create a model for this situation, but I can't seem to work out how to (easily) generate confidence and prediction intervals from it.
Would this be an appropriate model fit?
gls_1 <- gls(
response ~ group,
data = sample_df,
weights = varIdent(form = ~1|group)
)
Given this model, how would one generate the desired intervals?