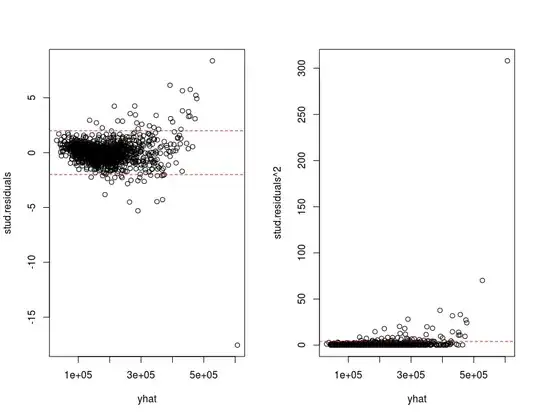
I have constructed a linear model (model3) and I'm trying to check the model's assumptions. As far the assumption of constant variance is concerned I have written the following piece of code
#constant variance
stud.residuals<-rstudent(model3)
yhat<-fitted(model3)
par(mfrow=c(1,2))
plot(yhat,stud.residuals)
abline(h=c(-2,2), col=2,lty=2)
plot(yhat, stud.residuals^2)
abline(h=4,col=2, lty=2)
ncvTest(model3)
yhat.quantiles<-cut(yhat, breaks=quantile(yhat, prbs=seq(0,1,0.25)), dig.lab=6)
table(yhat.quantiles)
leveneTest(rstudent(model3)~yhat.quantiles)
The output of the above is:
> ncvTest(model3)
Non-constant Variance Score Test
Variance formula: ~ fitted.values
Chisquare = 2999.046 Df = 1 p = 0
> table(yhat.quantiles)
yhat.quantiles
(30917.2,128460] (128460,167611] (167611,216345]
374 375 375
(216345,607215]
375
> leveneTest(rstudent(model3)~yhat.quantiles)
Levene's Test for Homogeneity of Variance (center = median)
Df F value Pr(>F)
group 3 52.424 < 2.2e-16 ***
1493
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
I'm confused about how to interpret p=0 from ncvTest. At first I thought it would be a matter of representation but ncvTest(model3)$p gives also zero.