@vucko gave me excellent answer on my question, unfortunately using Mathematica code. I'm trying to rewrite it in R and I'm lost in R functions providing kernel density estimation.
I have bivariate dataset with more than 46,000 rows (so I am also looking for a high performance solution--@vucko's solution is very time consuming). I would like to apply kernel density estimation and decide if some point lies in area with some density estimation level (confidence level respectively).
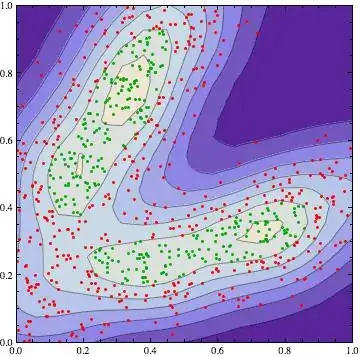
@vucko in his answer selected two groups. I need only to know if some point lies in the green group or not. And that should be done with R.
I experimented with kde and bkde2D functions but they don't provide me desired functionality as Mathematica SmoothKernelDistribution.
Can you please me show the direction? For normal distribution I found the ellipse function which approximated data with some confidence level and used inside.owin function.