I have a data frame called mxDF looks like this:
> head(mxDF)
y x.Age x.Weight x.Height x.Medicine x.genotype1
1 3.00 36 72 171 0 0
2 1.50 63 49 162 0 0
3 2.25 40 44 154 0 0
4 2.25 57 41 159 0 0
5 2.25 59 55 157 0 0
6 1.50 45 51 160 0 0
x.genotype2
1 0
2 0
Then I did the following:
mxMod <- glm(mxDF)
res.mxMod <- resid(mxMod);
plot(res.mxMod ~ mxDF[,1]);
abline(glm(res.mxMod ~ mxDF[,1]))
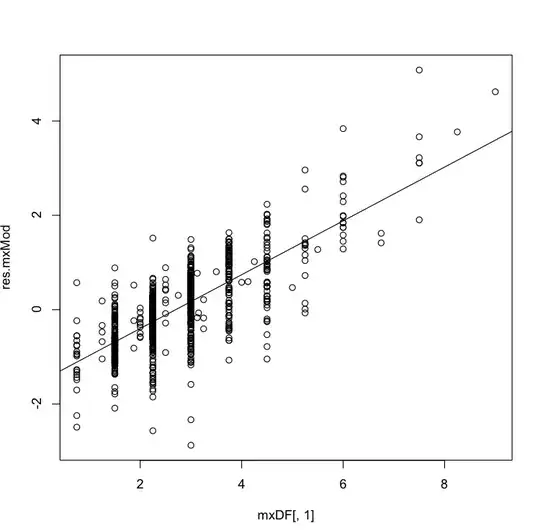
And the plot looks like below:
The pattern on the residue plot is so obvious, and then I did a glm on the residue ~ outcome:
summary(glm(res.mxMod ~ mxDF[,1]))
Call:
glm(formula = res.mxMod ~ mxDF[, 1])
Deviance Residuals:
Min 1Q Median 3Q Max
-3.04479 -0.28613 0.03926 0.36334 2.34211
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -1.54024 0.04782 -32.21 <2e-16 ***
mxDF[, 1] 0.57049 0.01630 34.99 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for gaussian family taken to be 0.3229295)
Null deviance: 693.21 on 923 degrees of freedom
Residual deviance: 297.74 on 922 degrees of freedom
AIC: 1581.8
Number of Fisher Scoring iterations: 2
Isn't this weird? What did I do wrong?