I have some data as shown below:
subject QM emotion yi
s1 75.1017 neutral -75.928276
s2 -47.3512 neutral -178.295990
s3 -68.9016 neutral -134.753906
s4 -193.6777 neutral -137.988266
s5 -89.8085 neutral 357.732239
s6 151.6949 neutral -342.555511
s7 -66.7561 neutral -55.791832
s8 176.7803 neutral 1.458443
s9 35.2962 neutral -196.741882
s10 -93.8680 neutral 11.772915
s11 22.2184 neutral 224.331467
s12 -57.7316 neutral -33.035969
s13 -24.6816 neutral -8.723662
s14 37.4021 neutral -66.085762
s15 -8.3483 neutral 87.531853
s16 66.6684 neutral -98.155365
s17 85.9628 neutral 2.060247
s1 17.2099 negative -104.168312
s2 -53.1114 negative -182.373474
s3 -33.0322 negative -137.420410
s4 -210.9767 negative -144.443954
s5 -19.8764 negative 408.798706
s6 112.5601 negative -375.723450
s7 -138.0572 negative -68.359787
s8 204.2617 negative -9.281775
s9 79.2678 negative -197.376678
s10 -135.2844 negative -9.184753
s11 -62.4264 negative 180.171204
s12 -77.7733 negative -28.135830
s13 57.8981 negative 20.544859
s14 11.4957 negative -65.592827
s15 18.5995 negative 124.318390
s16 122.0963 negative -76.976395
s17 107.1490 negative -28.010180
When I perform the following model with the nlme package in R:
mydata <- read.table('clipboard', header=T)
summary(lme(yi~QM, random=~1|subject, data=mydata))
I see a positive QM effect, which is also statistically significant at the 0.05 level:
Fixed effects: yi ~ QM
Value Std.Error DF t-value p-value
...
QM 0.36209 0.08492 16 4.263942 0.0006
Similar results for the QM effect can be obtained with two other models:
summary(lme(yi~QM*emotion, random=~1|subject, data=mydata))
summary(lme(yi~QM*emotion, random=~0+emotion|subject, data=mydata))
However, the result from a linear model tells a different story (negative QM effect, which is not statistically significant at the 0.05 level):
summary(lm(yi~QM, data=aggregate(mydata[,c('yi', 'QM')], by=list(mydata$subject), mean)))
Coefficients:
Estimate Std. Error t value Pr(>|t|)
...
QM -0.2973 0.4252 -0.699 0.495
Same things for two more linear models for each emotion separately:
summary(lm(yi ~ QM, data = mydata[mydata$emotion=='negative',]))
Coefficients:
Estimate Std. Error t value Pr(>|t|)
...
QM -0.1766 0.4055 -0.436 0.669
summary(lm(yi ~ QM, data = mydata[mydata$emotion=='neutral',]))
Coefficients:
Estimate Std. Error t value Pr(>|t|)
...
QM -0.3584 0.4251 -0.843 0.412
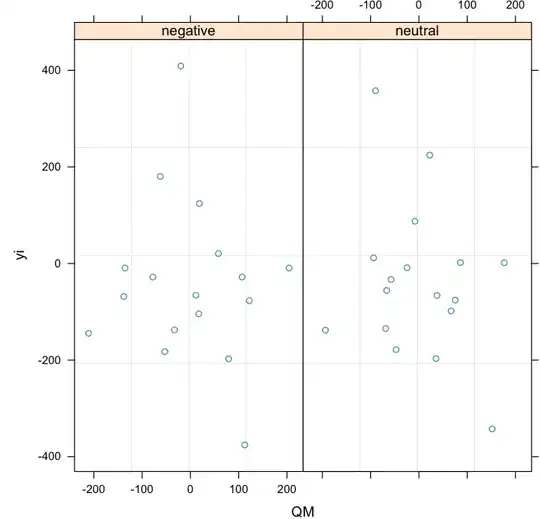
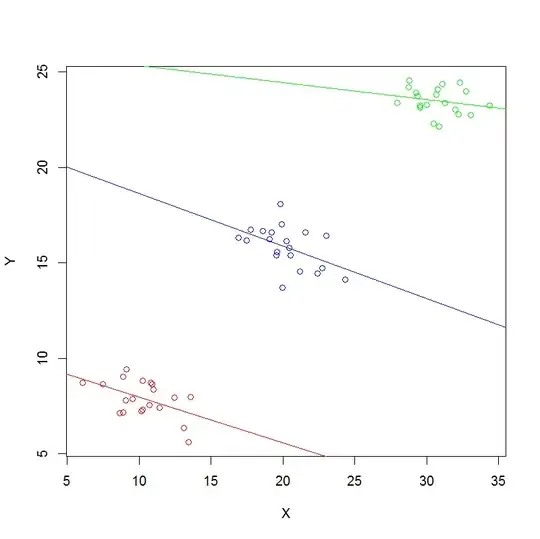
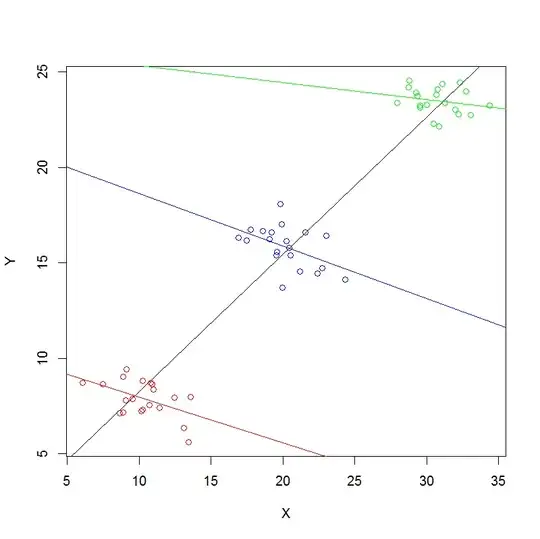
Admittedly the data is messy (see the attached plot). But still, what is causing the different assessment between lme() and lm()?