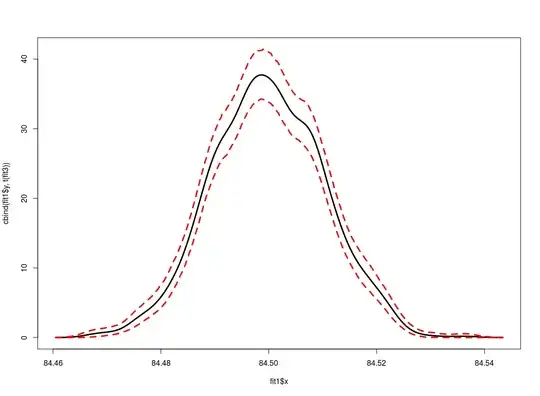
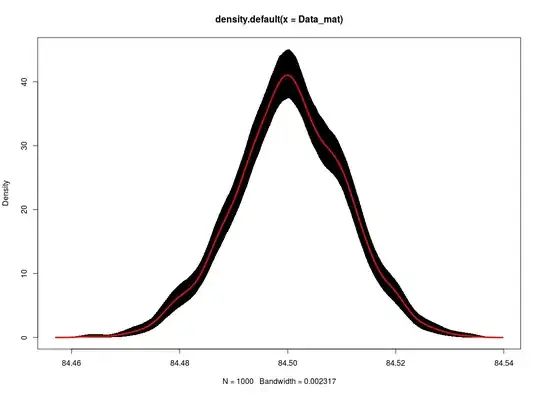
I found this excellent code snippet online which gives the code for boostrapping a kernel density estimate to get confidence bands. Now, I am not that well versed in R, and would like to know what's happening. I have commented in the source code below what I think each line does, but I might be wrong. Suggestions?
mu = 84.5; s = 0.01
Data_mat = rnorm(1000,mu,s)
#Generate Simple Kernel Density Estimate uing the default R function
fit1 <- density(Data_mat)
#Bootstrap starts
fit2 <- replicate(1000,
{
#Sample with replacement, for the bootstrap from the
#original dataset and save the resample to x
x <- sample(Data_mat, replace=TRUE)
#Generate the density from the resampled dataset, and
#extract y coordinates to generate variablity bands
#for that particular x coordinate in the smooth curve
density(x, from=min(fit1$x), to=max(fit1$x))$y
})
#Apply the quantile function to the y coordinates to get the
#bounds of the polygon to be drawn on the y axis?
#if so, why the 2.5% to 97.5% range? Am I missing a convention here?
fit3 <- apply(fit2, 1, quantile, c(0.025,0.975) )
#Adjust plot function to display line and variablity band from fit3
plot(fit1, ylim=range(fit3),xlim = c(-5,150))
#draw the actual polygon using the x coordinates from the original density
#and the y coordinates from calculated quantile variablity bands
polygon( c(fit1$x, rev(fit1$x)), c(fit3[1,], rev(fit3[2,])),
col='black', density = -0.5, border=F)
#Display the line again
lines(fit1)