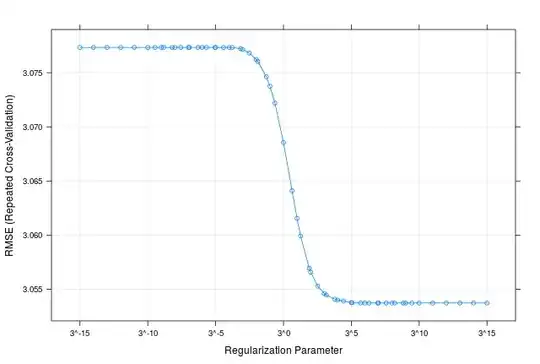
I am trying to do L2-regularized MLR on a data set using caret. Following is what I have done so far to achieve this:
r_squared <- function ( pred, actual){
mean_actual = mean (actual)
ss_e = sum ((pred - actual )^2)
ss_total = sum ((actual-mean_actual)^2 )
r_squared = 1 - (ss_e/ss_total)
}
df = as.data.frame(matrix(rnorm(10000, 10, 3), 1000))
colnames(df)[1] = "response"
set.seed(753)
inTraining <- createDataPartition(df[["response"]], p = .75, list = FALSE)
training <- df[inTraining,]
testing <- df[-inTraining,]
testing_response <- base::subset(testing,
select = c(paste ("response")))
gridsearch_for_lambda = data.frame (alpha = 0,
lambda = c (2^c(-15:15), 3^c(-15:15)))
regression_formula = as.formula (paste ("response", "~ ", " .", sep = " "))
train_control = trainControl (method="cv", number =10,
savePredictions =TRUE , allowParallel = FALSE )
model = train (regression_formula,
data = training,
trControl = train_control,
method = "glmnet",
tuneGrid =gridsearch_for_lambda,
preProcess = NULL
)
prediction = predict (model, newdata = testing)
testing_response[["predicted"]] = prediction
r_sq = round (r_squared(testing_response[["predicted"]],
testing_response[["response"]] ),3)
Here I am concerned about assurance that the model I am using for prediction is the best one (the optimal tuned lambda value).
P.S.: The data is sampled from random normal distribution, which is not giving a good R^2 value, but I want to get the idea correctly