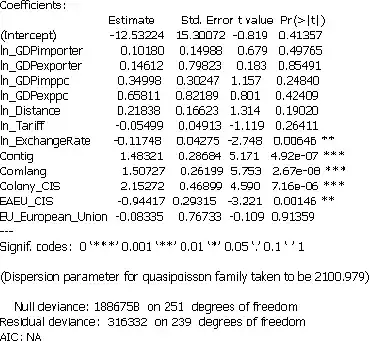
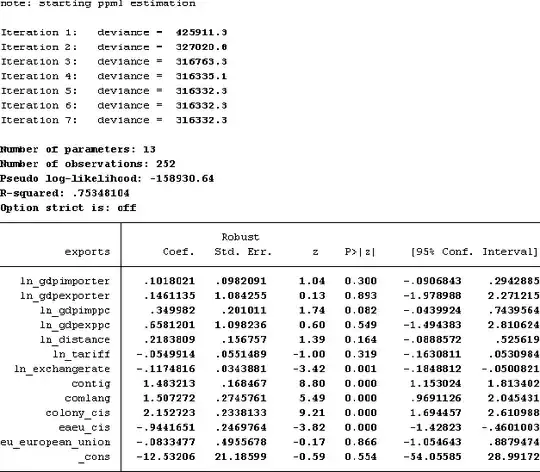
To expand on Wayne's excellent answer, ppml uses a robust (to heteroskedasticity) variance-covariance matrix and also a finite sample adjustment to that matrix to reduce bias.
These are very similar to what sandwich() from the package of the same name computes in R. The only difference is how the finite-sample adjustment is done. In the sandwich(...) function, no finite-sample adjustment is done at all by default, i.e., the sandwich is divided by 1/n where n is the number of observations. Alternatively, sandwich(..., adjust = TRUE) can be used which divides by 1/(n - k) where k is the number of regressors. Stata, however, divides by 1/(n - 1).
Here's how you can get R to match Stata by using a custom sandwich variance with an adjustment factor of 1/(n-1):
. clear
. set more off
. capture ssc install rsource
. use http://personal.lse.ac.uk/tenreyro/mock, clear
. saveold ~/Desktop/mock, version(12) replace
(saving in Stata 12 format, which can be read by Stata 11 or 12)
file ~/Desktop/mock.dta saved
. rsource, terminator(XXX) rpath("/usr/local/bin/R") roptions("--vanilla")
Assumed R program path: "/usr/local/bin/R"
Loading required package: zoo
Attaching package: 'zoo'
The following objects are masked from 'package:base':
as.Date, as.Date.numeric
Beginning of R output
R version 3.2.4 (2016-03-10) -- "Very Secure Dishes"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-apple-darwin13.4.0 (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
Natural language support but running in an English locale
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library("foreign")
> library("sandwich")
> library("lmtest")
> mock<-read.dta("~/Desktop/mock.dta")
> glmm<-glm(formula=y ~ x + w, family=quasipoisson(link="log"),data=mock)
>
> sandwich1 <- function(object, ...) sandwich(object) * nobs(object) / (nobs(object) - 1)
> coeftest(glmm,vcov=sandwich1)
z test of coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.516969 0.098062 5.2718 1.351e-07 ***
x 0.125657 0.101591 1.2369 0.2161
w 0.013410 0.710752 0.0189 0.9849
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
>
End of R output
.
. ppml y x w
note: checking the existence of the estimates
Number of regressors excluded to ensure that the estimates exist: 0
Number of observations excluded: 0
note: starting ppml estimation
note: y has noninteger values
Iteration 1: deviance = 139.7855
Iteration 2: deviance = 137.7284
Iteration 3: deviance = 137.7222
Iteration 4: deviance = 137.7222
Number of parameters: 3
Number of observations: 100
Pseudo log-likelihood: -173.89764
R-squared: .01628639
Option strict is: off
------------------------------------------------------------------------------
| Robust
y | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
x | .1256565 .1015913 1.24 0.216 -.0734588 .3247718
w | .0134101 .7107518 0.02 0.985 -1.379638 1.406458
_cons | .5169689 .0980624 5.27 0.000 .3247702 .7091676
------------------------------------------------------------------------------
Here's the Stata/R code that generates the output above. I am using rsource to run R from Stata (and you will need to tweak the rpath() below to match your setup), but that is not really necessary: you can just run the rsource part from R.
clear
set more off
capture ssc install rsource
use http://personal.lse.ac.uk/tenreyro/mock, clear
saveold ~/Desktop/mock, version(12) replace
rsource, terminator(XXX) rpath("/usr/local/bin/R") roptions("--vanilla")
library("foreign")
library("sandwich")
library("lmtest")
mock<-read.dta("~/Desktop/mock.dta")
glmm<-glm(formula=y ~ x + w, family=quasipoisson(link="log"),data=mock)
sandwich1 <- function(object, ...) sandwich(object) * nobs(object) / (nobs(object) - 1)
coeftest(glmm,vcov=sandwich1)
XXX
ppml y x w