I have some data with more variables than observations, that I'd like to subject to a principal components analysis. For didactic reasons (to give an intuition for factor retention criteria under parallel analysis), I am here interested in the distribution of the residual correlations.
Let this be my data (sampled from real data):
data <- c(1,-1,-3,-1,1,1,-2,-2,1,3,-3,0,2,4,0,0,-1,0,-1,-4,-2,3,-2,2,0,4,-3,-1,0,2,2,-4,0,3,0,1,-2,-1,-3,2,-1,4,-4,0,3,2,-3,-2,4,-1,3,0,1,0,-3,2,1,-2,1,-1,1,1,-4,3,2,0,-2,0,0,0,-1,0,-3,3,-4,2,0,-1,0,0,1,1,0,-3,1,-3,4,-2,0,-1,-4,-1,-2,2,2,-2,0,1,0,2,-1,3,3,-1,4,0,-2,1,-4,0,-3,-2,-2,-1,-1,-3,1,3,1,-3,2,-2,2,3,0,-1,-4,4,-1,1,0,0,3,2,-1,0,0,0,2,-2,4,1,0,1,-1,-2,-3,-1,1,-2,-2,-1,0,-1,-4,1,2,2,0,0,1,3,4,0,-4,-1,4,0,-2,3,0,3,-3,0,2,1,2,-3,1,0,-3,0,-4,1,1,2,0,3,0,-1,1,-1,2,-2,1,3,0,0,-3,-3,-4,-1,4,-2,3,2,-2,-1,-1,2,0,1,0,0,-2,4,1,4,0,0,1,-1,-4,-1,2,0,-3,0,3,-1,1,-1,-4,4,-3,-2,1,-2,0,-3,3,-1,-2,2,0,0,-2,3,2,0,1,2,1,-2,-2,1,-3,1,-4,0,3,3,-1,-1,3,1,0,2,2,-2,-4,-3,0,-1,2,0,2,0,-2,-1,0,-1,1,0,0,4,-3,4,0,4,-1,-2,3,-4,0,1,-2,-2,0,-1,1,0,-4,-2,-3,0,-1,1,-3,2,0,0,-1,3,0,2,2,-3,3,2,4,-1,1,1,-1,3,-2,-2,1,0,-3,-2,0,-1,0,-4,4,-3,3,1,2,2,-2,0,0,0,-1,0,1,-1,-4,3,-3,2,1,1,4,0,-1,2,-2,-1,0,2,0,3,-3,-4,0,-1,0,-3,2,2,-1,-1,0,2,-4,0,0,1,-2,1,3,0,-3,4,-2,4,-1,1,3,-2,1,1,-1,1,-4,-1,-2,1,-3,-3,2,2,-4,-3,-1,-2,0,2,2,1,0,1,3,0,0,-2,4,1,-2,-1,0,0,4,3,0,0,-1,3,2,2,1,4,1,3,-3,3,-2,-1,0,0,0,-1,1,0,2,-1,-4,-2,-4,1,-1,4,0,-2,-3,3,0,1,-2,-1,0,2,-3,0,2,2,-4,-2,-2,2,-1,0,1,1,-3,-3,4,1,1,3,3,0,-4,-1,-3,-2,3,1,4,2,-2,0,0,0,0,0,0,-1,-1,-1,0,0,-1,4,-1,2,-2,-2,3,-1,-4,-2,1,-3,-2,2,2,4,-4,-1,0,-1,1,3,1,1,-3,0,-3,2,0,0,1,0,0,3,4,2,0,1,1,0,-4,1,4,0,0,-3,2,-3,-1,3,0,0,3,-1,-4,-1,-1,2,3,-1,-2,-2,0,2,-3,-2,1,0,-2,1,4,0,-1,-2,0,0,-3,-1,0,-3,-2,0,3,-4,0,1,0,3,1,-2,-1,1,-1,3,4,-2,-4,2,-3,2,2,-1,1,1,0,2,-3,0,-2,-2,0,0,-3,0,1,3,-1,-1,2,0,1,0,-1,3,-4,-3,-2,3,-2,2,4,0,-4,4,-1,2,1,2,0,1,-1,1,3,3,-2,3,0,1,-3,4,2,-2,0,-4,1,-1,0,2,-1,2,0,0,-4,0,4,-1,-2,1,-3,1,0,2,-1,-1,-3,0,-2,1,-1,0,-1,0,0,3,-4,-3,0,1,-3,-2,4,-4,1,2,4,0,-2,-2,-1,0,-1,1,3,0,-3,0,-1,1,2,2,1,2,-2,3,-4,1,0,-3,-1,-2,-1,-2,4,1,0,2,3,-2,-3,0,-1,3,0,0,1,4,-4,0,2,-3,0,0,-2,-1,1,-1,3,2,2,1,-2,1,-2,0,-3,1,-4,0,1,3,-1,-3,0,-2,0,-1,1,0,-2,0,0,-1,-1,-4,0,4,-3,1,-1,2,2,3,4,2,3,2,4,3,-2,4,0,3,-4,-3,3,-1,-1,-3,2,-2,1,2,1,2,-4,-1,-2,-1,-3,2,1,1,-2,0,-1,1,0,0,0,0,0,0,-3,4,-4,3,-1,0,-3)
data <- c(data, 2,3,-1,0,-2,3,1,-1,1,1,2,-1,1,-4,0,-1,0,2,2,-3,1,-2,0,-2,0,4,0,-2,0,-2,4,-3,0,-2,0,-1,-4,0,3,-1,-3,1,2,4,0,1,0,-1,-2,-1,3,-3,0,1,3,-4,2,-2,0,1,-1,2,2,0,1,-4,0,-3,0,4,-2,-3,-1,3,-2,-2,3,2,2,2,3,1,1,-4,1,-2,2,0,1,1,0,-3,0,0,-1,-1,-1,-1,0,0,4,-1,-2,-2,-1,-2,0,-3,-1,4,-1,-2,0,4,-3,0,0,-1,3,-4,1,0,0,1,0,2,1,-4,3,3,2,2,2,1,0,-3,1,-1,3,-2,0,0,2,-4,0,0,2,2,0,0,-1,-1,1,-2,3,-4,-1,1,-1,-3,2,-2,4,-3,0,1,3,0,4,1,1,-3,-2,-2,2,-2,0,-2,1,-3,-3,0,2,-4,0,4,1,0,0,4,3,-4,3,0,-1,2,-1,1,1,-3,0,-2,0,-1,2,1,-1,3,-1,0,2,-4,1,0,3,-2,-1,0,2,1,-3,0,-4,3,-1,0,0,-3,-2,-2,1,3,-1,0,2,-3,2,0,4,-1,-2,4,1,-1,1,0,3,0,-1,-1,2,-3,-1,-2,0,-2,-2,0,-3,3,0,0,3,-4,-1,0,2,1,-2,-1,1,-4,4,-3,2,1,0,4,1,2,1,1,-2,-1,-3,-1,1,0,3,4,0,-2,-4,-2,-2,0,0,1,4,-4,0,2,3,-3,2,2,-1,0,-1,2,1,-1,3,0,1,-3,0,-2,4,-1,-1,-2,-2,-3,1,3,0,-1,-4,4,0,0,3,-1,2,-2,-1,0,-3,2,-3,3,1,-4,1,0,1,0,1,2,0,0,2,2,0,-3,-2,1,1,-3,-1,-1,2,0,-1,2,0,0,0,2,4,-4,-2,0,-2,0,1,3,3,-4,1,-1,3,-3,0,1,-1,-2,4,-4,-3,-3,2,1,-1,-1,-2,4,1,0,-2,-2,1,3,-1,0,4,-4,0,0,0,3,3,-1,0,-3,2,0,-1,1,-2,2,2,0,1,3,3,-3,-2,-1,0,-3,-1,-2,2,-4,-2,3,-2,4,0,0,2,-1,-1,0,2,0,0,2,1,-4,4,-1,1,0,1,1,0,-3,1,-2,1,-3,-1,-1,1,-2,1,-1,2,-2,-3,2,-4,-1,0,3,3,3,0,-4,0,4,-3,0,4,-2,2,2,0,0,0,0,1,-1,1,-2,1,-3,-1,-1,1,-3,-1,2,-1,-2,-2,2,-4,-1,1,2,1,-3,3,0,0,4,-2,0,1,-4,0,3,0,4,2,3,0,0,0,3,4,-3,4,-3,-2,-3,3,-2,2,0,2,3,0,0,2,2,1,-4,0,0,0,1,-1,1,1,-4,-1,-2,1,0,0,-1,-1,-2,-1,-3,1,0,3,-3,1,-1,3,2,-1,-4,-2,3,4,-1,0,-2,4,0,0,-1,1,2,1,2,2,-4,-1,-2,1,0,0,0,-2,-3,0,4,3,0,-1,0,3,-4,1,1,-1,0,-3,2,4,2,3,0,2,-2,-3,-1,1,2,-4,0,1,-3,1,-2,-1,-2,0,-1,0,-2,0,-2,2,-2,-3,0,2,-4,0,-1,1,0,-3,1,-3,1,0,2,4,-4,-2,-1,1,-2,0,1,3,0,3,0,0,-1,-1,3,4,-1,2,-1,1,-1,-3,0,2,-1,-2,2,4,-1,-4,3,-2,3,0,3,-1,-3,-2,0,2,0,-4,-3,4,-2,2,1,1,1,0,1,0,0,0,0,3,-3,-2,-1,1,-1,1,3,-1,-1,-4,1,0,0,2,-2,-3,-4,0,-3,4,1,0,1,0,-1,3,-2,0,-2,0,4,2,2,2,0,1,-2,-4,1,0,-4,0,4,-1,-3,-3,2,-3,0,1,2,2,-2,-1,4,-1,3,-1,3,0,-2,0,0,1,-2,1,2,-1,0,3,-2,-1,-1,0,0,3,0,0,1,2,-2,-2,2,0,1,3,2,4,-3,0,-1,1,-3,-4,0,3,-4,-2,0,2,-3,1,-1,1,-1,4,-4,-3,-3,-1,0,-1,-2,0,1,-1,1)
data <- c(data, -2,0,0,1,3,2,4,-2,-3,-1,0,0,3,0,2,-1,-2,2,2,0,1,-4,4,3,1,0,2,-4,0,0,-2,-3,-1,1,0,-2,-1,3,1,-2,-1,3,2,-4,-1,-1,2,4,0,3,0,-3,2,-2,1,1,1,4,0,-3,0,0,4,-2,-2,0,1,-4,4,0,0,-4,-2,1,-2,2,2,3,-1,-3,-1,0,0,3,0,2,-1,-3,1,-3,1,-1,2,3,-1,0,1,0,4,-2,-2,0,0,-2,3,-1,-1,3,2,0,-2,4,-1,-1,0,-4,-1,0,1,0,-3,1,1,-4,2,-3,0,-3,1,3,2,2,1)
data <- matrix(data = data, nrow = 36)
(sorry about the clunky input).
Now, let's look at the initial correlation and then the residual correlations.
(I'm doing this here with psych::principal for the sake of convenience, but I've also calculated the residuals by hand with prcomp, with same results).
cor <- cor(data, method="pearson") # figure out initial cors
library(psych)
pca <- principal(r = data, nfactors = 8, residuals = TRUE, rotate = "none") # I know 8 factors is ridiculous, but that's the didactiv point I'm trying to make.
#> Warning in cor.smooth(r): Matrix was not positive definite, smoothing was
#> done
#> In factor.scores, the correlation matrix is singular, an approximation is used
#> Warning in cor.smooth(r): Matrix was not positive definite, smoothing was
#> done
# Calculate residuals ===
cor <- as.matrix(cor) # just to be sure
loa <- pca$loadings # take just the loas
res <- NULL
res <- array(data = NA, dim = c(nrow(cor), ncol(cor), ncol(loa)), dimnames = list(cor = NULL, cor = NULL, PC = 1:ncol(loa))) # this is what the residuals array should look lile
for (i in 1:ncol(loa)) {
if (i == 1) {
res[,, i] <- cor - loa[, i] %*% t(loa[, i])
} else {
res[,, i] <- res[,, i-1] - loa[, i] %*% t(loa[, i])
}
}
# we now have an array with var, var, PC as dimensions
# Take only upper triangle (without diagonal)
take.tri <- function(cors) { # make this little helper function
cors <- cors[upper.tri(x = cors, diag = FALSE)]
return(cors)
}
res.df <- apply(X = res, MARGIN = 3, FUN = take.tri)
# add the original correlation as 0th PC =======
res.df <- cbind(`0` = take.tri(cor), res.df)
# make plot =====
library(reshape2)
library(ggplot2)
res.df <- melt(data = res.df)
colnames(res.df) <- c("Obs", "PC", "Cor")
g <- NULL
g <- ggplot(data = res.df, mapping = aes(x = Cor, color = PC, group = PC))
g <- g + geom_freqpoly(binwidth = 0.05)
g <- g + xlim(-1,1)
g
#> Warning: Removed 18 rows containing missing values (geom_path).
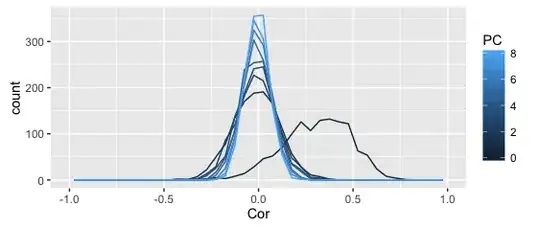
The above plots shows the distribution of correlation coefficients (denoted as PC 0), as well as the residual correlations from the first 8 principal component, all in one frequency polygon plot.
The smoother plot uses the full dataset, the rough one just the above sample (can't share the full dataset).
I get why the distribution becomes more leptopkurtic (aka: steep) as more components are extracted; that's the whole point of PCA (and this illustration): the remaining correlation matrices approximate a singular matrix with all zeros (and 1s on the diagonal).
I also understand why the original correlations are asymmetric -- given my kind of data (Q-sorts), that frequently happens.
What I don't understand is why the asymmetry seems to disappear completely after the first PC is extracted. Shouldn't the asymmetry dissipate slowly as more PCs are extracted?
My questions are:
- Is this to be expected?
- Is this in the logic of PCA, or a computational artefact?
- Is this approach meaningful to illustrate parallel analysis / why you shouldn't overretain components?
Also, as a bonus, I'd be really curious what the expected distribution of correlation coefficients from random data would be, but that's another question.