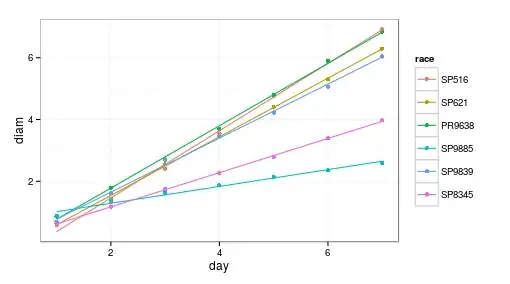
It depends on what you mean by statistically different. Statistically different from each other?
Looking at your plot, you've got four that look pretty clearly the same and then two that are much different. So, if you run:
library(lme4)
summary(lmer(diam~day*race + (1+day|race), data=long))
You get, in part:
Fixed effects:
Estimate Std. Error t value
(Intercept) -0.71786 0.11409 -6.292
day 1.08902 0.11022 9.880
raceSP621 0.36143 0.16135 2.240
racePR9638 0.48036 0.16135 2.977
raceSP9885 1.46143 0.16135 9.057
raceSP9839 0.61643 0.16135 3.820
raceSP8345 0.78071 0.16135 4.839
day:raceSP621 -0.13982 0.15588 -0.897
day:racePR9638 -0.07982 0.15588 -0.512
day:raceSP9885 -0.81652 0.15588 -5.238
day:raceSP9839 -0.21429 0.15588 -1.375
day:raceSP8345 -0.53491 0.15588 -3.432
lmer doesn't give p-values, because calculating degrees of freedom for these models isn't entirely straightforward, but looking at the t-value, you can see that you've got big values (in absolute terms) for the day:SP9885 interaction and the day:SP8345 interaction. This suggests that the slopes for those two conditions are shallower than the slopes for the others.
Technically, this is treating the SP516 group as the baseline, and testing everything else for differences from that.
If you wanted to set a different group as the baseline, you could run:
long$race <- relevel(long$race, ref='SP9885')
summary(lmer(diam~day*race + (1+day|race), data=long))
Truncated output:
Fixed effects:
Estimate Std. Error t value
(Intercept) 0.7436 0.1176 6.324
day 0.2725 0.1086 2.508
raceSP516 -1.4614 0.1663 -8.789
raceSP621 -1.1000 0.1663 -6.615
racePR9638 -0.9811 0.1663 -5.900
raceSP9839 -0.8450 0.1663 -5.082
raceSP8345 -0.6807 0.1663 -4.094
day:raceSP516 0.8165 0.1536 5.314
day:raceSP621 0.6767 0.1536 4.404
day:racePR9638 0.7367 0.1536 4.795
day:raceSP9839 0.6022 0.1536 3.920
day:raceSP8345 0.2816 0.1536 1.833
If you're jonesin' for a p-value, you can see this faq
EDIT:
Using multilevel model here because I'm assuming that the observations across days are not independent for each of the fungi races. Thus, you've nested data.