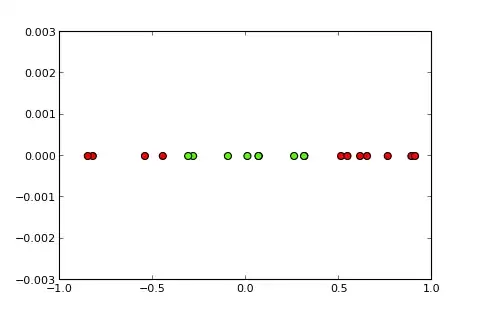
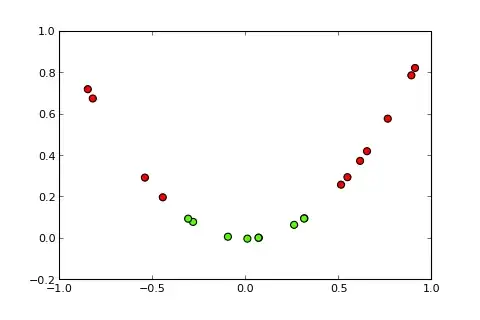
Classification problems with nonlinear boundaries cannot be solved by a simple perceptron. The following R code is for illustrative purposes and is based on this example in Python):
nonlin <- function(x, deriv = F) {
if (deriv) x*(1-x)
else 1/(1+exp(-x))
}
X <- matrix(c(-3,1,
-2,1,
-1,1,
0,1,
1,1,
2,1,
3,1), ncol=2, byrow=T)
y <- c(0,0,1,1,1,0,0)
syn0 <- runif(2,-1,1)
for (iter in 1:100000) {
l1 <- nonlin(X %*% syn0)
l1_error <- y - l1
l1_delta <- l1_error * nonlin(l1,T)
syn0 <- syn0 + t(X) %*% l1_delta
}
print("Output After Training:")
## [1] "Output After Training:"
round(l1,3)
## [,1]
## [1,] 0.488
## [2,] 0.468
## [3,] 0.449
## [4,] 0.429
## [5,] 0.410
## [6,] 0.391
## [7,] 0.373
Now the idea of a kernel and the so-called kernel trick is to project the input space into a higher dimensional space, like so (sources of pics):
My question
How do I make use of the kernel trick (e.g. with a simple quadratic kernel) so that I get a kernel perceptron, which is able to solve the given classification problem? Please note: This is mainly a conceptual question but if you could also give the necessary code modification this would be great
What I tried so far
I tried the following which works alright but I think that this is not the real deal because it becomes computationally too expensive for more complex problems (the "trick" behind the "kernel trick" is not just the idea of a kernel itself but that you don't have to calculate the projection for all instances):
X <- matrix(c(-3,9,1,
-2,4,1,
-1,1,1,
0,0,1,
1,1,1,
2,4,1,
3,9,1), ncol=3, byrow=T)
y <- c(0,0,1,1,1,0,0)
syn0 <- runif(3,-1,1)
Full Disclosure
I posted this question a week ago on SO but it didn't get much attention. I suspect that here is a better place because it is more a conceptual question than a programming question.