I got a time series dataset from the lab and would like to fit my data to a curve (using R package): $$ P_{1}(t)=y_{0}-a_{1}e^{-b_{1}t}-a_{2}e^{-b_{2}t}, b_{2}>b_{1} \tag{A} $$
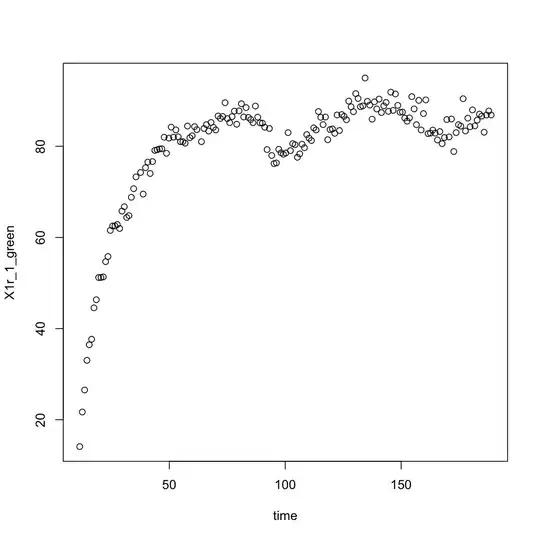
I plotted the data:
I think I need to start with good initial values to get a better estimate when fitting to the curve.So I start with a simpler curve: $$ P_{2}(t)=a_{1}e^{-b_{1}t} \tag{B} $$
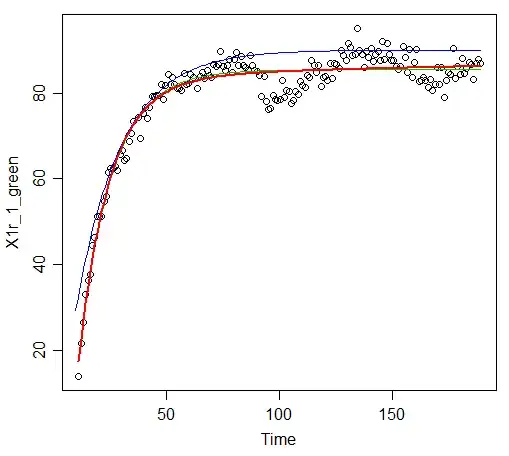
and I got:
Call:
lm(formula = log(P1_1) ~ time)
Coefficients:
(Intercept) time
4.084148 0.002703
That is, $a_{1}=54, b_{1}=0.003$ (roughly).
So I set the starting value $y_{0}=30, a_{1}=20, b_{1}=0.002, b_{1}=30, b_{2}=0.003$ and then did:
nls_fit <- nls(X1r_1_green~-y0+A1*exp(tau1*Time)+A2*exp(tau2*Time),
start=list(y0=35, A1=20, A2=30, tau1=.02, tau2=.03), data=data1, trace=T)
However, it returns errors that I could not figure out how to fix, such as:
step factor 0.000488281 reduced below 'minFactor' of 0.000976562
singular gradient matrix at initial parameter estimates
After digging into some posts online, the error might be the poor initial values. After many times attempts at different initial values I've still had no luck, though. Do you have any suggestions?
The data is as follows:
Time X1r_1_green
1 11.333 14.1060
2 12.443 21.6894
3 13.450 26.5231
4 14.457 33.0356
5 15.463 36.4517
6 16.471 37.6585
7 17.478 44.5417
8 18.485 46.3301
9 19.492 51.2111
10 20.499 51.2094
11 21.505 51.3405
12 22.513 54.6848
13 23.520 55.8172
14 24.527 61.5436
15 25.534 62.5158
16 26.541 62.5279
17 27.547 62.8395
18 28.555 61.9790
19 29.562 65.7806
20 30.569 66.7353
21 31.576 64.3837
22 32.583 64.7834
23 33.589 68.8058
24 34.597 70.6975
25 35.604 73.2809
26 37.597 74.2578
27 38.707 69.5155
28 39.714 75.3023
29 40.721 76.5360
30 41.727 74.0348
31 42.735 76.6561
32 43.742 79.1025
33 44.749 79.2463
34 45.756 79.4334
35 46.763 79.4644
36 47.769 81.9903
37 48.776 78.4627
38 49.784 81.7913
39 50.791 84.2073
40 51.798 82.0027
41 52.805 83.6003
42 53.811 82.0862
43 54.818 80.9991
44 55.826 81.0088
45 56.833 80.6877
46 57.840 84.4431
47 58.847 81.8827
48 59.853 82.3165
49 60.861 84.3172
50 61.868 83.6584
51 63.862 81.0175
52 64.972 83.9112
53 65.979 84.7511
54 66.986 83.3383
55 67.993 85.2289
56 68.999 84.1657
57 70.007 83.5959
58 71.014 86.6437
59 72.021 86.1192
60 73.028 86.5437
61 74.034 89.5566
62 75.041 86.0854
63 76.049 85.1923
64 77.056 86.5120
65 78.063 87.7273
66 79.070 84.8408
67 80.076 87.7706
68 81.083 89.3330
69 82.091 86.4411
70 83.098 88.4944
71 84.105 86.3283
72 85.112 85.8619
73 86.118 85.1758
74 87.125 88.8349
75 88.133 86.4028
76 89.140 85.1487
77 90.147 85.0808
78 91.154 84.1891
79 92.160 79.2782
80 93.167 83.9142
81 94.175 77.9845
82 95.182 76.1993
83 96.189 76.3565
84 97.196 79.3567
85 98.202 78.5809
86 99.210 78.2514
87 100.217 78.5353
88 101.224 82.9957
89 102.231 79.0445
90 103.237 80.5854
91 104.244 80.3490
92 105.252 77.5888
93 106.259 78.3460
94 107.266 80.4607
95 108.273 79.6868
96 109.279 82.5708
97 110.287 81.7843
98 111.294 81.2645
99 112.301 84.0419
100 113.308 83.5896
101 114.315 87.6291
102 115.321 86.3707
103 116.329 84.7563
104 117.336 86.4180
105 118.343 81.4442
106 119.350 83.6793
107 120.357 83.8090
108 121.363 82.8488
109 122.371 86.8810
110 123.378 83.4640
111 124.385 86.9706
112 125.392 86.5779
113 126.398 85.8226
114 127.406 89.9185
115 128.413 88.6702
116 129.420 87.5920
117 130.427 91.5657
118 131.434 90.5003
119 132.440 88.6852
120 133.448 88.8704
121 134.455 94.9899
122 135.462 89.8719
123 136.469 89.0472
124 137.477 85.9579
125 138.483 89.7549
126 139.490 88.1925
127 140.497 90.3598
128 141.504 87.4038
129 142.511 88.8794
130 143.518 89.6318
131 144.525 87.6712
132 145.532 91.8890
133 146.539 87.8800
134 147.546 91.4838
135 148.553 88.9816
136 149.560 87.4804
137 150.567 87.5171
138 151.574 86.1898
139 152.581 85.5146
140 153.588 86.2422
141 154.595 90.9029
142 155.602 88.2005
143 156.609 84.7370
144 157.616 90.0859
145 158.623 83.5787
146 159.630 87.1796
147 160.637 90.1887
148 161.644 82.7800
149 162.651 82.8887
150 163.658 83.5638
151 164.665 82.8934
152 165.672 81.3959
153 166.679 83.2340
154 167.686 80.5834
155 168.693 81.9224
156 169.700 85.8687
157 170.707 82.0637
158 171.714 85.9917
159 172.722 78.8532
160 173.728 83.0037
161 174.735 84.7516
162 175.742 84.4288
163 176.749 90.4443
164 177.757 83.3640
165 178.764 86.1808
166 179.770 84.2887
167 180.777 87.9906
168 181.784 84.4754
169 182.791 85.7398
170 183.798 87.0640
171 184.805 86.6323
172 185.812 83.0898
173 186.819 86.8609
174 187.826 87.7454
175 188.833 86.8763