I suggest to use package deming. Here is an example of Deming regression using the package:
library(deming)
fit.lm <- lm(aes ~ aas, data=arsenate)
fit.deming <- deming(aes ~ aas, data=arsenate, stdpat = c(1, 0, 1, 0))
print(fit.deming)
#Call:
#deming(formula = aes ~ aas, data = arsenate, stdpat = c(1, 0, 1, 0))
#
#n= 30
# Coef se(coef) lower 0.95 upper 0.95
#Intercept 0.4293665 0.3079456 -0.1741959 1.032929
#Slope 0.8759571 0.1180054 0.6446708 1.107244
#
# Scale= 0.7899665
fit.errors <- deming(aes ~ aas, data=arsenate, xstd=se.aas, ystd=se.aes)
print(fit.errors)
#Call:
#deming(formula = aes ~ aas, data = arsenate, xstd = se.aas, ystd = se.aes)
#
#n= 30
# Coef se(coef) lower 0.95 upper 0.95
#Intercept 0.1064481 0.2477071 -0.3790489 0.5919451
#Slope 0.9729928 0.1429651 0.6927863 1.2531993
#
# Scale= 1.165495
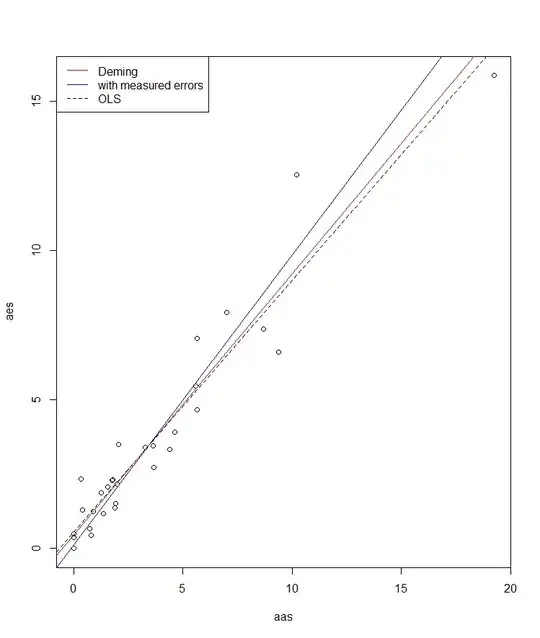
plot(aes ~ aas, data=arsenate)
abline(fit.lm, lty = 2)
abline(fit.deming, col = "red")
abline(fit.errors, col = "blue")
legend("topleft", legend = c("Deming", "with measured errors","OLS"),
lty = c(1, 1:2), col = c("red", "blue","black"))
Apparently the Jackknife technique is used to get the standard errors. You probably should read the package vignette and have a look at the references therein.
