This is probably an embarrassingly easy question, but where else can I turn to...
I'm trying to put together examples of regression with mixed effects using lmer {lme4}, so that I can present [R] code that automatically downloads toy datasets in Google Drive and run every instance in this blockbuster post.
And starting with the first case (i.e. V1 ~ (1|V2) + V3, where V3 is a continuous variable acting as a fixed effect, and V2 is Subjects, both trying to account for V1, a continuous DV), I was expecting to retrieve different intercepts for each one of the Subjects and a single slope for all of them. Yet, this was not the case consistently.
I don't want to bore you with the origin or meaning of the datasets below, because I'm sure most of you get the idea without much explaining. So let me show you what I get... If you're so inclined you can just copy and paste in [R]... it should work if you have {lme4} in your Environment:
Expected Output:
politeness <- read.csv("http://www.bodowinter.com/tutorial/politeness_data.csv")
head(politeness)
subject gender scenario attitude frequency
1 F1 F 1 pol 213.3
2 F1 F 1 inf 204.5
3 F1 F 2 pol 285.1
4 F1 F 2 inf 259.7
library(lme4)
fit <- lmer(frequency ~ (1|subject) + attitude, data = politeness)
coefficients(fit)
$subject
(Intercept) attitudepol
F1 241.1352 -19.37584
F2 266.8920 -19.37584
F3 259.5540 -19.37584
M3 179.0262 -19.37584
M4 155.6906 -19.37584
M7 113.2306 -19.37584
Surprising Output:
library(gsheet)
recall <- read.csv(text =
gsheet2text('https://drive.google.com/open?id=1iVDJ_g3MjhxLhyyLHGd4PhYhsYW7Ob0JmaJP8MarWXU',
format ='csv'))
head(recall)
Subject Time Emtl_Value Recall_Rate Caffeine_Intake
1 Jim 0 Negative 54 95
2 Jim 0 Neutral 56 86
3 Jim 0 Positive 90 180
4 Jim 1 Negative 26 200
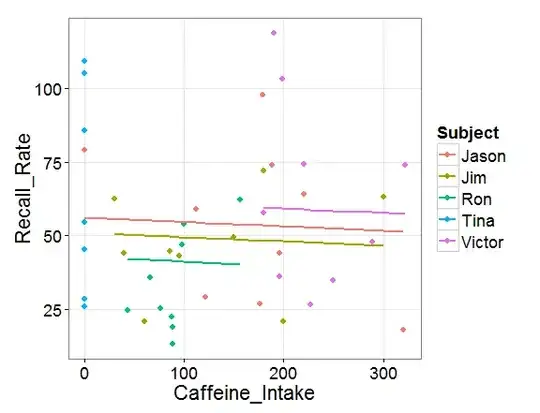
fit <- lmer(Recall_Rate ~ (1|Subject) + Caffeine_Intake, data = recall)
coefficients(fit)
$Subject
(Intercept) Caffeine_Intake
Jason 51.51206 0.013369
Jim 51.51206 0.013369
Ron 51.51206 0.013369
Tina 51.51206 0.013369
Victor 51.51206 0.013369
Here is the output of (summary(fit)):
Linear mixed model fit by REML ['lmerMod']
Formula: Recall_Rate ~ (1 | Subject) + Caffeine_Intake
Data: recall
REML criterion at convergence: 413.9
Scaled residuals:
Min 1Q Median 3Q Max
-1.54125 -0.98422 0.04967 0.81465 1.83317
Random effects:
Groups Name Variance Std.Dev.
Subject (Intercept) 0.0 0.00
Residual 601.2 24.52
Number of obs: 45, groups: Subject, 5
Fixed effects:
Estimate Std. Error t value
(Intercept) 51.51206 5.92408 8.695
Caffeine_Intake 0.01337 0.03792 0.353
Correlation of Fixed Effects:
(Intr)
Caffen_Intk -0.787
Question:
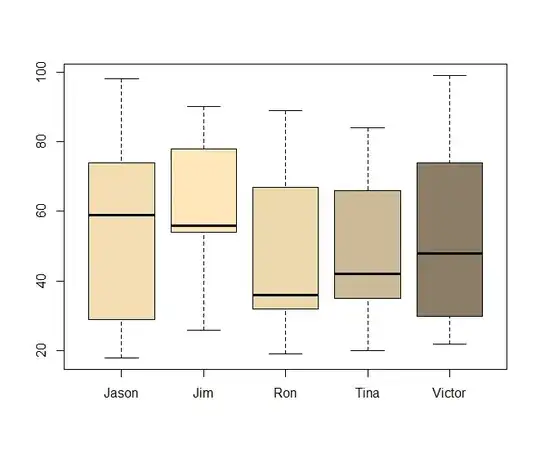
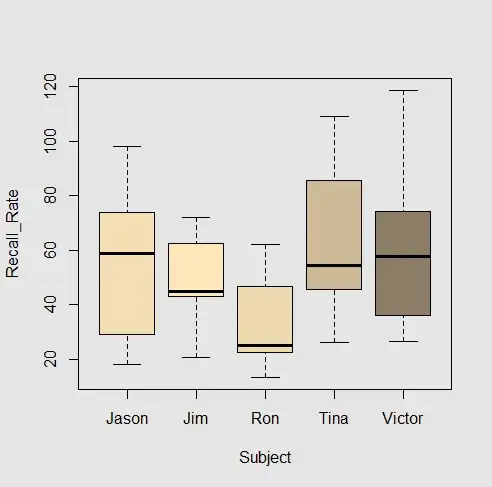
Why are all the Intercepts for the different subjects the same in the second example? The structure of the datasets and the lmer syntax appear very similar... and the boxplots don't seem to support the result:
Thank you in advance!