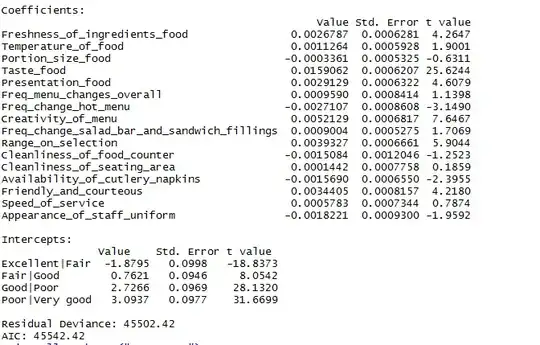
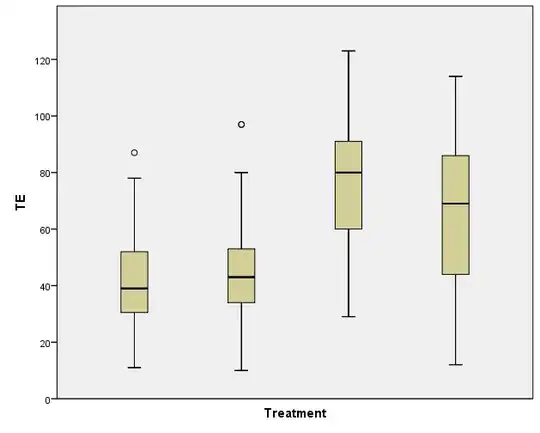
I'm using SPSS to run a GLM (general linear model) univariate with 1 fixed factor (Treatment) and one random factor (experimental replicate). There are 4 treatment groups. The measurement is number of cells per embryo. The Levene's Test for Equality of Error Variances is significant (P=0.000) and I can see from the Spread vs Level plot that there may be a pattern.
What are my options from here?
I have tried log transforming my data, and that increased the P value of the Levene's Test to P=0.02, but there still appears to be pattern in the Spread vs Level plot.
I know that I could use a Post hoc test that does not assume equal variances (Tamhane's T2 or Dunnett's T3), or I could use a Kruskal-Wallis H, but both of these are only possible with 1 factor, not two.
I would really appreciate any help with this!