Background: I've been given a dataframe that contains data for a CDF. The column X contains the 250 $X$ values, and the column P contains $p(X\geq x)$.
I paste the dataset below:
X <- c(59639.83,396409.6,733179.5,1069949,1406719,1743489,2080259,2417029,2753798,3090568,3427338,3764108,4100878,4437647,4774417,5111187,5447957,5784727,6121496,6458266,6795036,7131806,7468576,7805346,8142115,8478885,8815655,9152425,9489195,9825964,10162734,10499504,10836274,11173044,11509813,11846583,12183353,12520123,12856893,13193662,13530432,13867202,14203972,14540742,14877512,15214281,15551051,15887821,16224591,16561361,16898130,17234900,17571670,17908440,18245210,18581979,18918749,19255519,19592289,19929059,20265829,20602598,20939368,21276138,21612908,21949678,22286447,22623217,22959987,23296757,23633527,23970296,24307066,24643836,24980606,25317376,25654146,25990915,26327685,26664455,27001225,27337995,27674764,28011534,28348304,28685074,29021844,29358613,29695383,30032153,30368923,30705693,31042463,31379232,31716002,32052772,32389542,32726312,33063081,33399851,33736621,34073391,34410161,34746930,35083700,35420470,35757240,36094010,36430780,36767549,37104319,37441089,37777859,38114629,38451398,38788168,39124938,39461708,39798478,40135247,40472017,40808787,41145557,41482327,41819097,42155866,42492636,42829406,43166176,43502946,43839715,44176485,44513255,44850025,45186795,45523564,45860334,46197104,46533874,46870644,47207414,47544183,47880953,48217723,48554493,48891263,49228032,49564802,49901572,50238342,50575112,50911881,51248651,51585421,51922191,52258961,52595731,52932500,53269270,53606040,53942810,54279580,54616349,54953119,55289889,55626659,55963429,56300198,56636968,56973738,57310508,57647278,57984047,58320817,58657587,58994357,59331127,59667897,60004666,60341436,60678206,61014976,61351746,61688515,62025285,62362055,62698825,63035595,63372364,63709134,64045904,64382674,64719444,65056214,65392983,65729753,66066523,66403293,66740063,67076832,67413602,67750372,68087142,68423912,68760681,69097451,69434221,69770991,70107761,70444531,70781300,71118070,71454840,71791610,72128380,72465149,72801919,73138689,73475459,73812229,74148998,74485768,74822538,75159308,75496078,75832848,76169617,76506387,76843157,77179927,77516697,77853466,78190236,78527006,78863776,79200546,79537315,79874085,80210855,80547625,80884395,81221165,81557934,81894704,82231474,82568244,82905014,83241783,83578553,83915323)
P <- c(0.9496295,0.3940323,0.1819006,0.1004568,0.06435878,0.04517915,0.03321521,0.02513521,0.01949269,0.01578545,0.01260615,0.0104641,0.008522678,0.007116022,0.006081262,0.005251359,0.004476574,0.00385598,0.00340083,0.002963359,0.002529698,0.002248921,0.001873215,0.001684442,0.001561909,0.001383793,0.001255323,0.001091163,0.001016704,0.0008913803,0.0008493767,0.0008085694,0.0007478901,0.0006541131,0.0005959151,0.0005181331,0.0004730944,0.0004529344,0.0004312237,0.0003961764,0.0003765924,0.0003047255,0.0002763022,0.0002419195,0.0002233103,0.0002040808,0.0001989854,0.0001915639,0.0001906778,0.0001787147,0.0001592858,0.00015902,0.0001587984,0.0001469683,0.0001410089,0.0001282483,0.0001235296,0.0001094619,8.651059e-05,8.602321e-05,8.159245e-05,6.947432e-05,6.896478e-05,6.32491e-05,6.32491e-05,6.304971e-05,6.304971e-05,6.231864e-05,6.121095e-05,4.927005e-05,4.915928e-05,4.683313e-05,3.768361e-05,3.768361e-05,3.768361e-05,3.768361e-05,3.768361e-05,3.768361e-05,3.768361e-05,3.768361e-05,1.586212e-05,1.586212e-05,1.586212e-05,1.586212e-05,1.586212e-05,1.586212e-05,1.586212e-05,1.586212e-05,1.586212e-05,1.586212e-05,1.586212e-05,1.586212e-05,1.586212e-05,1.586212e-05,1.559628e-05,1.526397e-05,1.513105e-05,1.513105e-05,1.513105e-05,1.513105e-05,1.513105e-05,7.820291e-06,7.820291e-06,7.310754e-06,7.310754e-06,7.310754e-06,7.310754e-06,7.310754e-06,7.310754e-06,6.446756e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.677531e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06,3.234455e-06)
data <- data.frame(X,P)
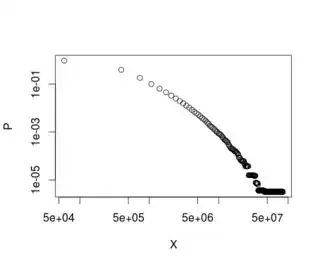
When I plot it in log-log scale, this is the result:
plot(data, log="xy")
Now, I've been asked by my boss to fit a Pareto distribution to this plot, so that a straight line appears. Pareto functions are like this: $f(x)=Ax^{-\alpha}$, and they sometimes have a minimum $x$ from where they fit empirical data.
Question: How could I fit the Pareto distribution to this data?
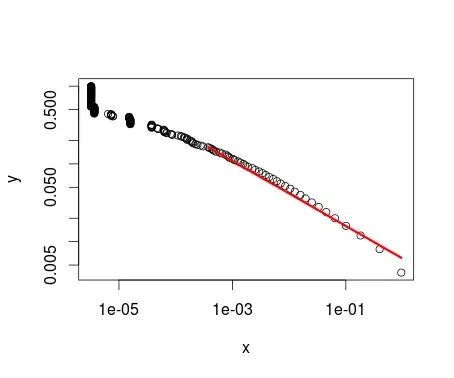
I haven't been able to do that. I've used the poweRlaw package using the $X$ column as input, but I'm not really sure that such a thing is statistically correct:
pareto <- conpl$new(data$P)
est <- estimate_xmin(pareto)
pareto$setXmin(est)
The resuting parameters are: $x_{min}=0.0003765924$ and $\alpha=1.418147$. The plot of this operation is the following:
plot(pareto)
lines(pareto, col=2, lwd=3)
Obviously, the $x_{min}$ doesn't refer to any value in $X$, but instead refers to a value present in $P$. And, as you can see, the shape of the distribution is quite the same as the other plot, but upside down.